dummy slide
Real Data Science USA - R Meetup
A Decade of Using R
in Production
Gergely Daróczi
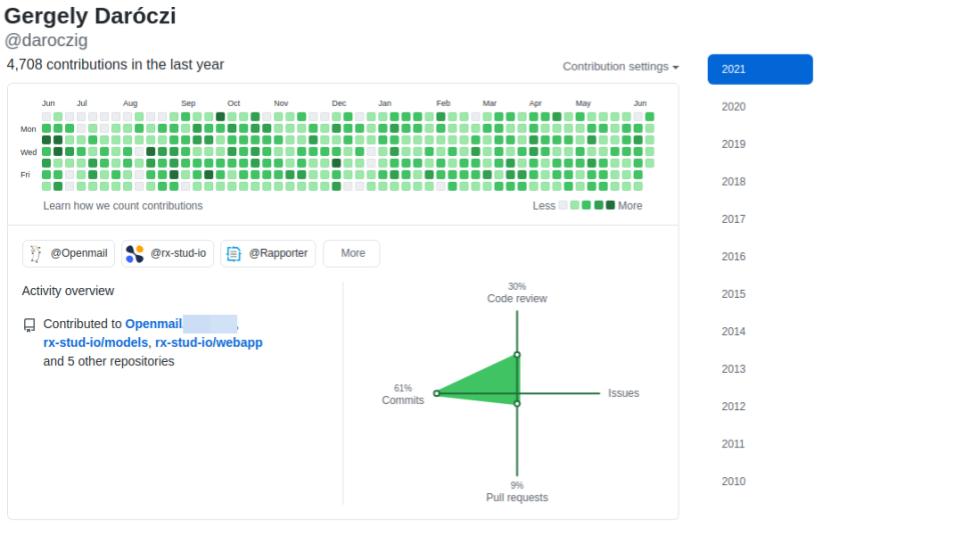
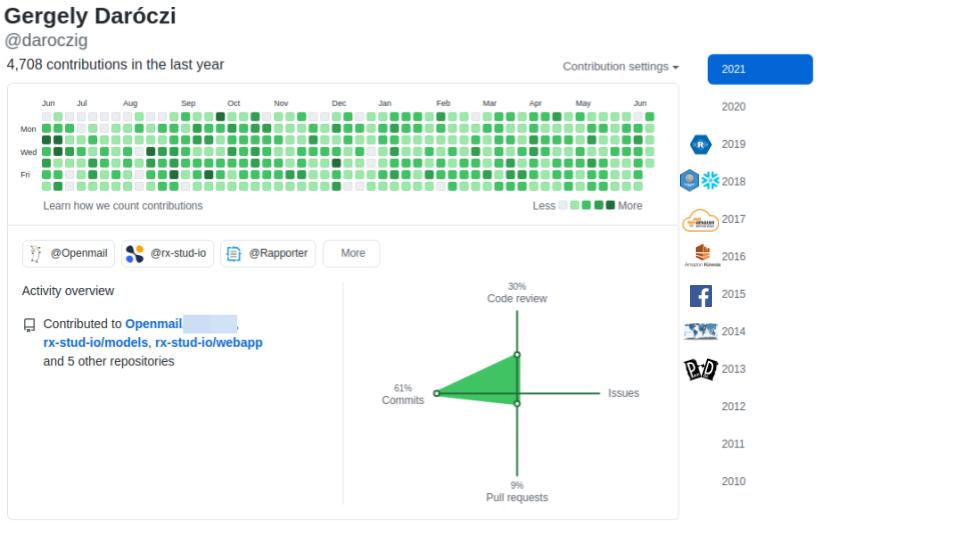
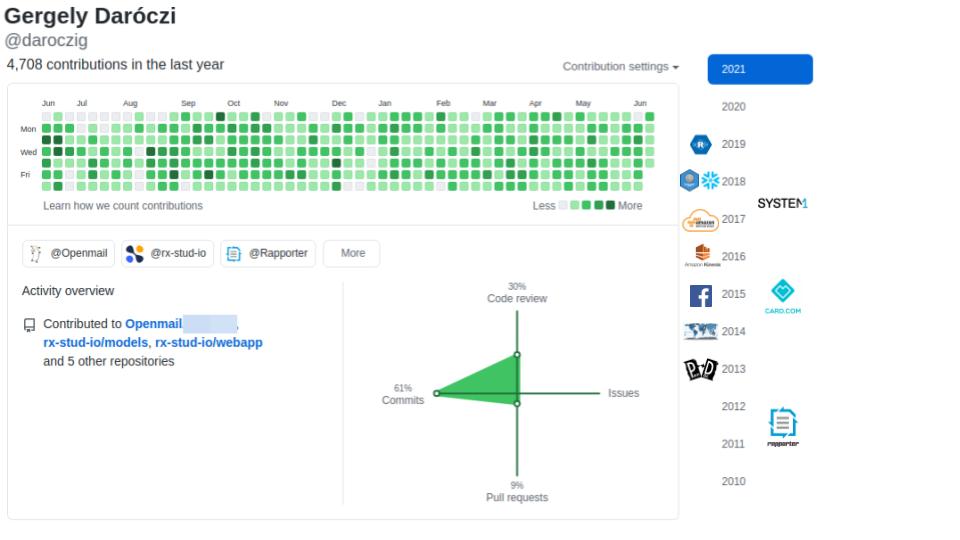
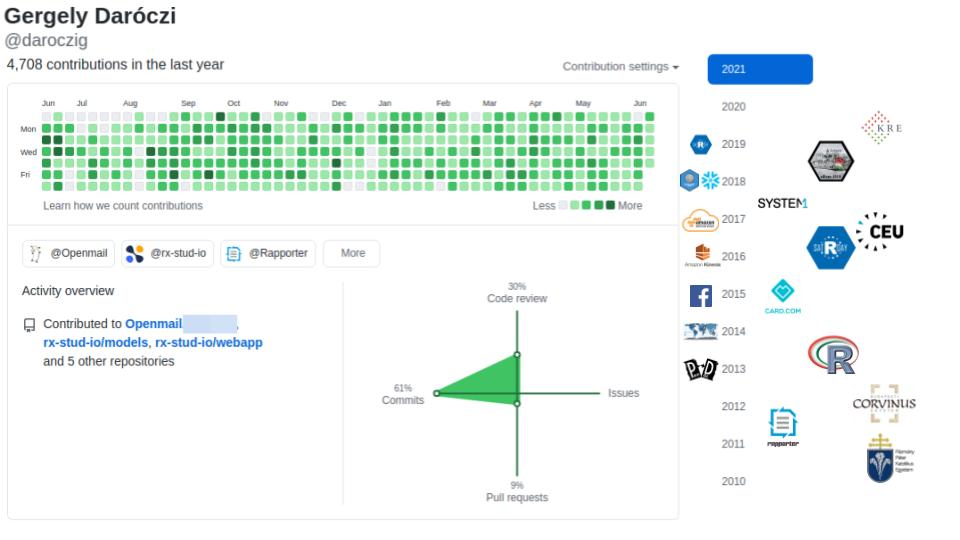
@daroczig
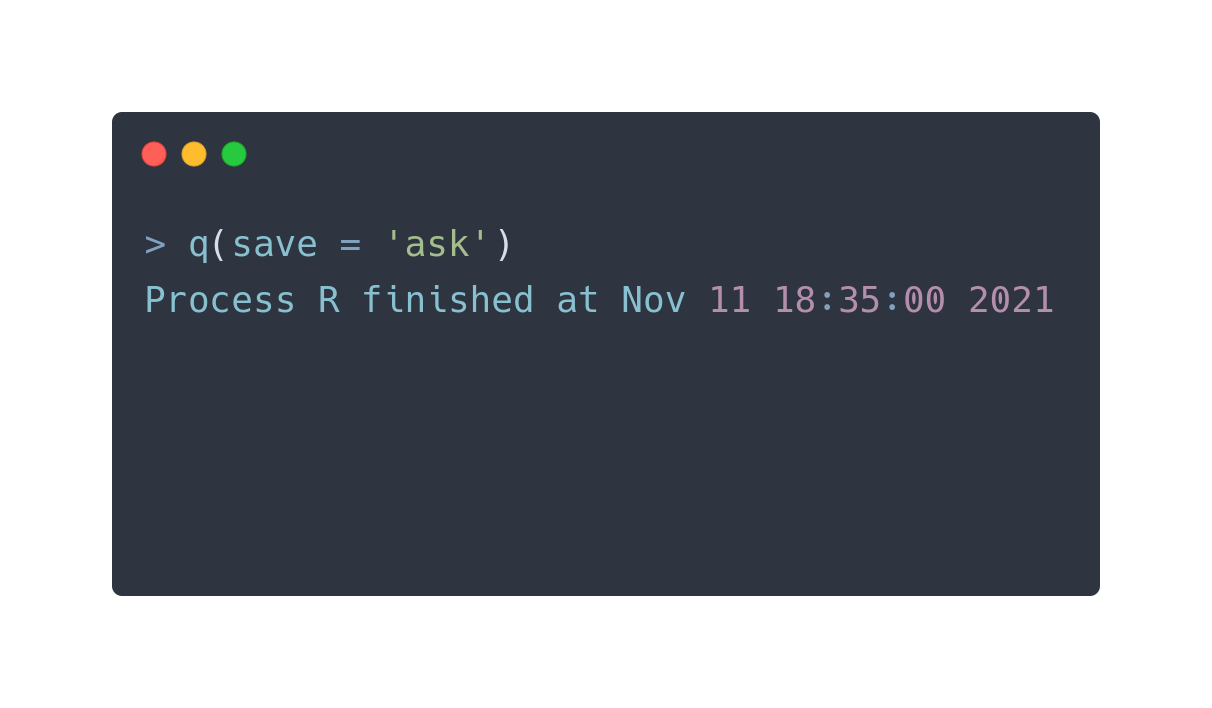
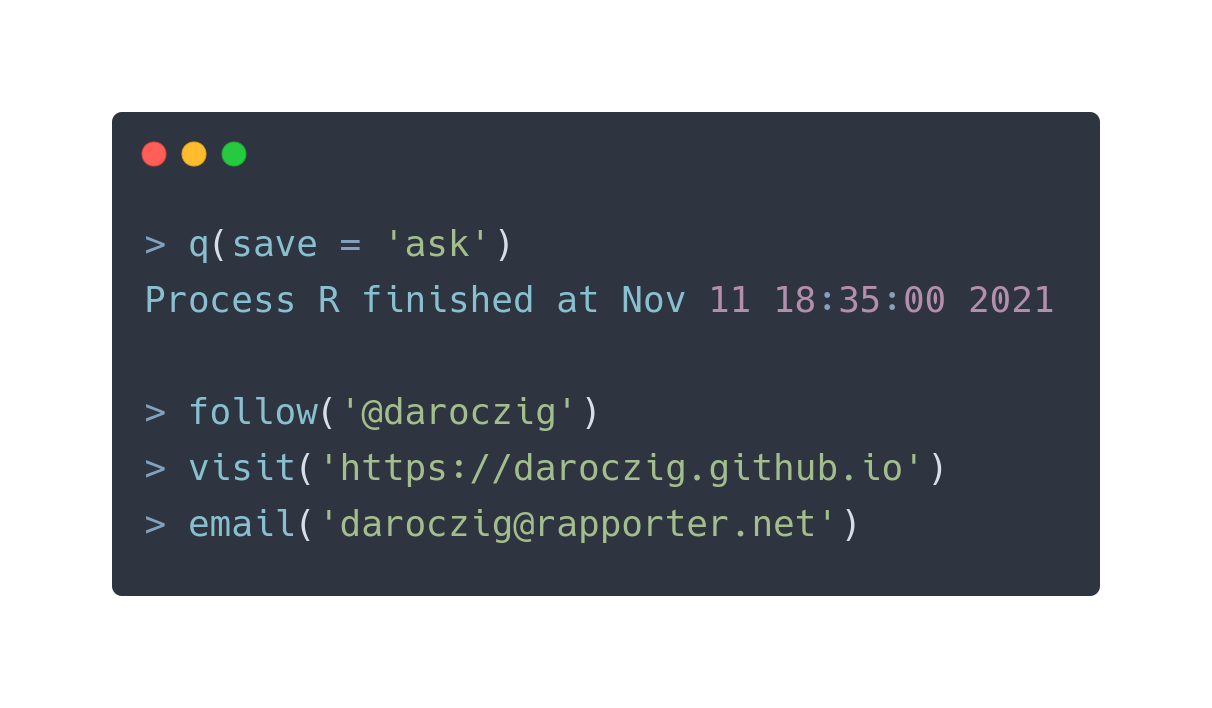
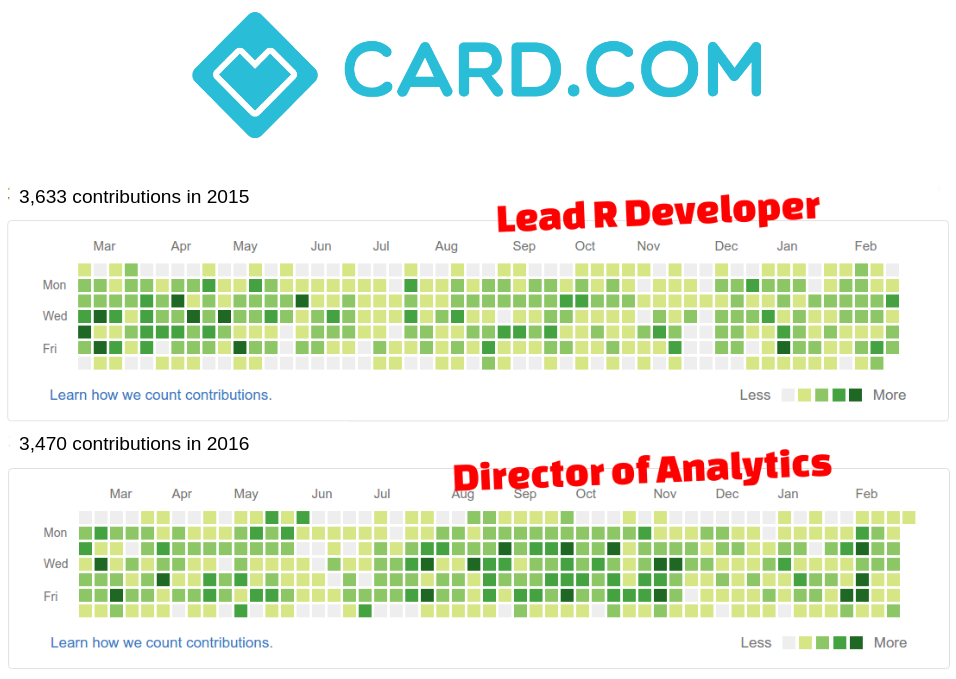
$ whoami
$ whoami
$ curl https://r.console
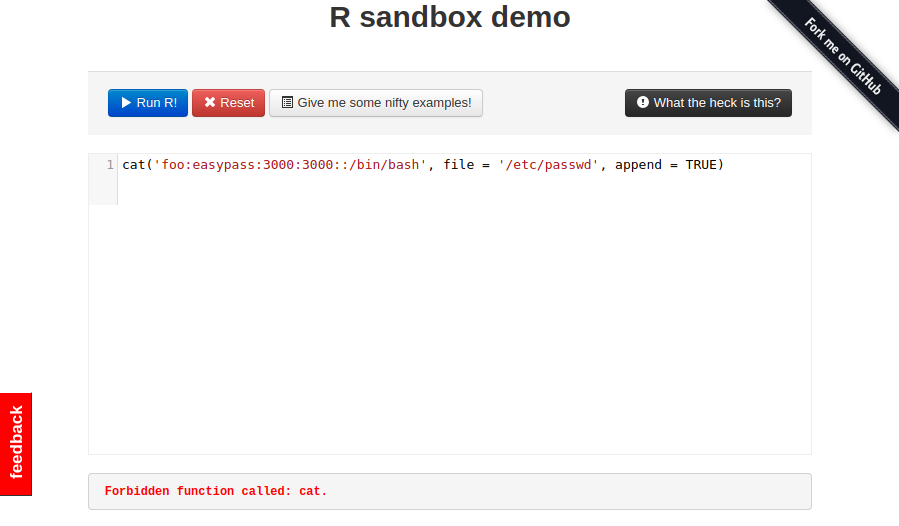
# cat /etc/passwd
$ whoami
$ whoami
$ whoami
$ whoami
$ whoami
> Sys.setenv(env = "prod")

> ??production
> ??production
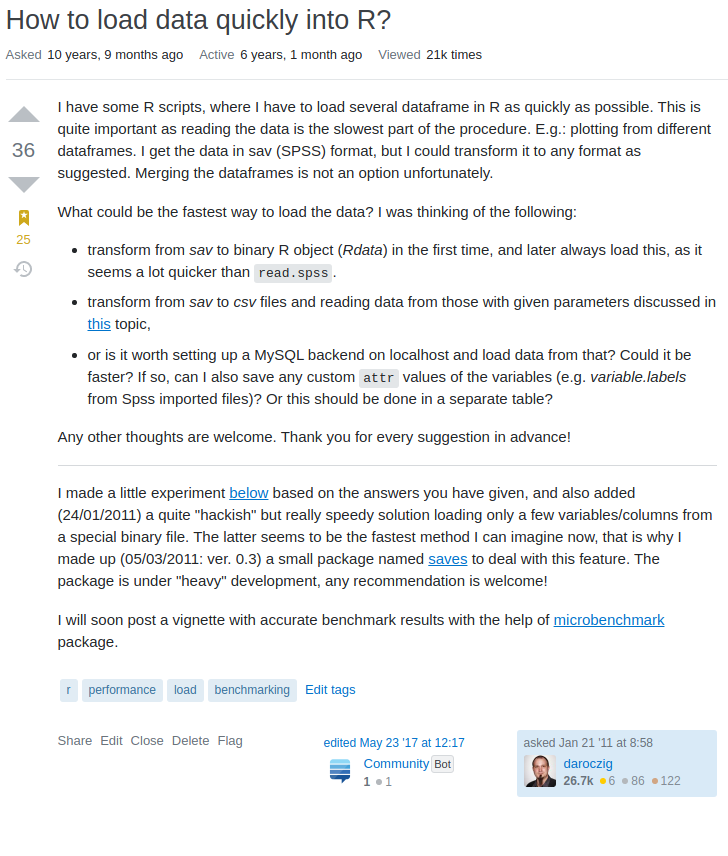
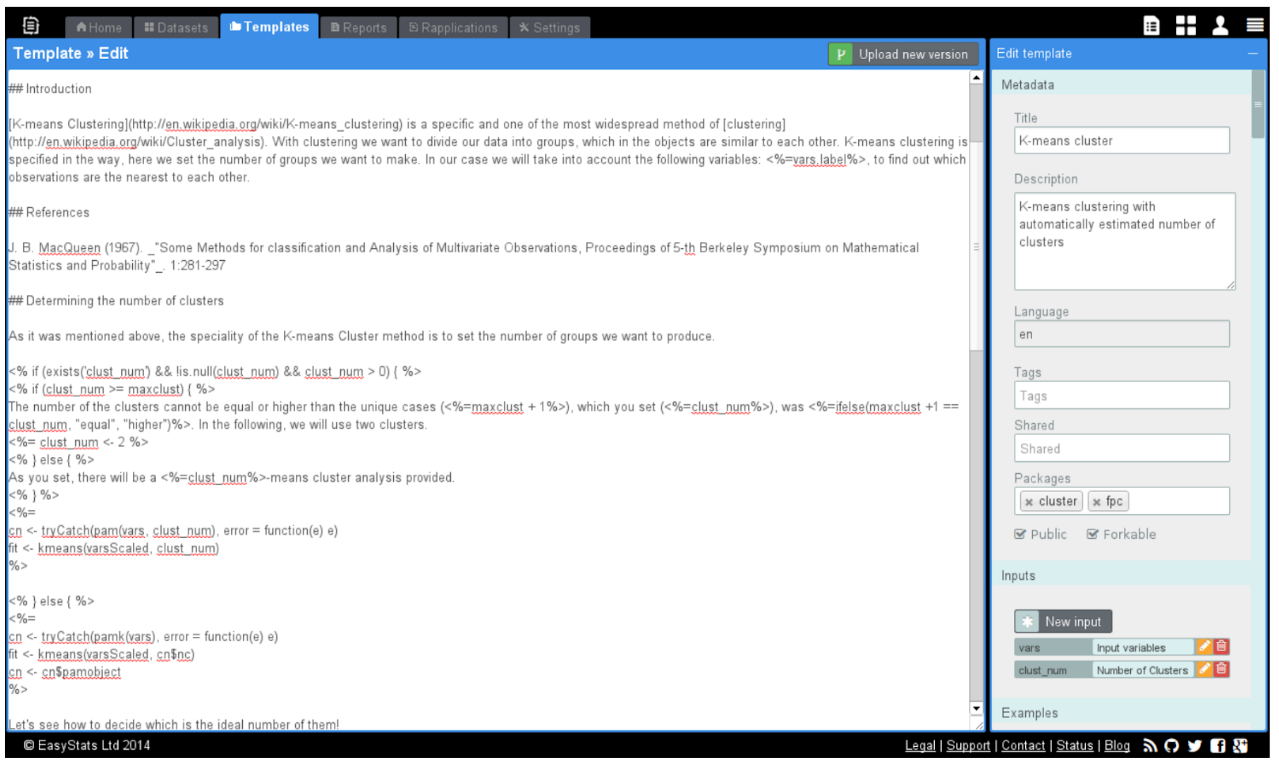
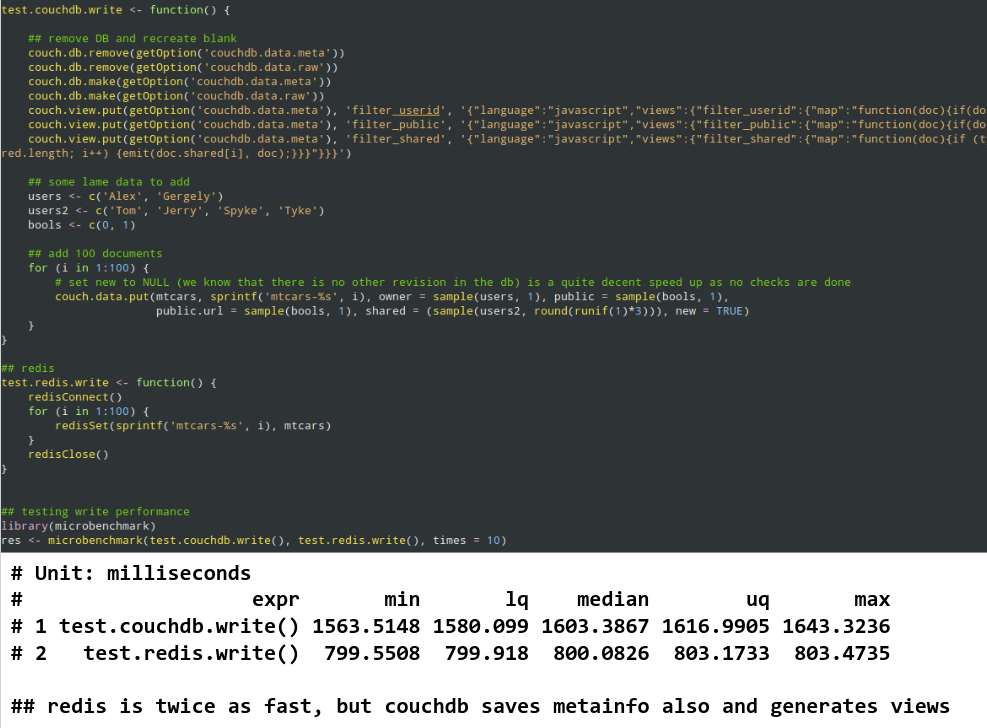
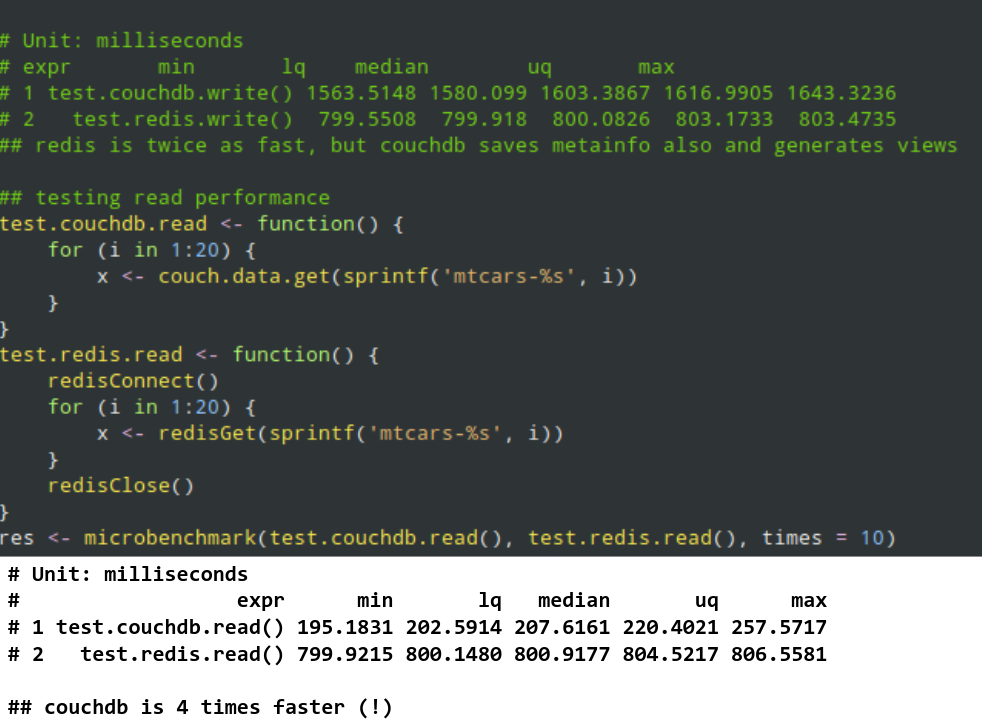
- 2006: Calling R scripts from PHP (both reading from MySQL) to generate custom plots embedded in a homepage
- 2008: Automated/batch R scripts to generate thousands of pages of crosstables, ANOVA and plots from SPSS with
pdflatex - 2011: Ruby on Rails web application with
RApacheandpandocto report in plain English (NoSQL databases, scaling, security, central error tracking etc) - 2012: Plain RApache web application for NLP and network analysis
- 2015: Standardizing the data infrastructure of a fintech startup to use R for reporting, batch jobs, and stream processing
- 2017: Redesign, monitor and scale the DS infrastructure of an adtech startup for batch training and live scoring
> production <<- list(…)
> production <<- list(…)
Using in R in a non-interactive way:
- Running R without manual intervention (e.g. scheduled via CRON, triggered via upstream job trigger or API request)
- Need for a standard, e.g. containerized environment (pinned R and package versions, OS packages,
.Rprofileetc) - Security! (e.g. safeguarded production environment, encrypted credentials, aware of Little Bobby Tables, AppArmor etc)
- Job output is informative (logging), recorded (logging) and monitored (e.g.
errorhandler for ErrBit, CloudWatch logs or Splunk etc), alerts and notifications
> isTRUE(interactive())
> traceback()
> tryCatch
- Use version control!
- Use CI/CD tools!
- Write clean code! DRY!
- Document! Open-source!
- Log everything! Snapshot everything!
- Security!
- Use a scalable job scheduler!
- Dockerize your environment!
- Pin your package versions!
Check out the “Productionizing R scripts in the cloud” at satRday LA 2019!
> sessionInfo()


> usethis::create_package

> ??log
> library(data.table)
> packages <- data.table(available.packages())
> ## avoid analog, logit, (archeo|bio|genea|hydro|topo|...)logy
> packages[grepl('(?<!ana)log(?![it|y])', Package, perl = TRUE), Package]
[1] "adjustedcranlogs" "bayesloglin" "blogdown"
[4] "CommunityCorrelogram" "cranlogs" "efflog"
[7] "eMLEloglin" "futile.logger" "gemlog"
[10] "gglogo" "ggseqlogo" "homologene"
[13] "lifelogr" "log4r" "logbin"
[16] "logconcens" "logcondens" "logcondens.mode"
[19] "logcondiscr" "logger" "logging"
[22] "loggit" "loggle" "logKDE"
[25] "loglognorm" "logmult" "lognorm"
[28] "logNormReg" "logOfGamma" "logspline"
[31] "lolog" "luzlogr" "md.log"
[34] "mdir.logrank" "mpmcorrelogram" "PhylogeneticEM"
[37] "phylogram" "plogr" "poilog"
[40] "rChoiceDialogs" "reactlog" "rmetalog"
[43] "robustloggamma" "rsyslog" "shinylogs"
[46] "ssrm.logmer" "svDialogs" "svDialogstcltk"
[49] "tabulog" "tidylog" "wavScalogram"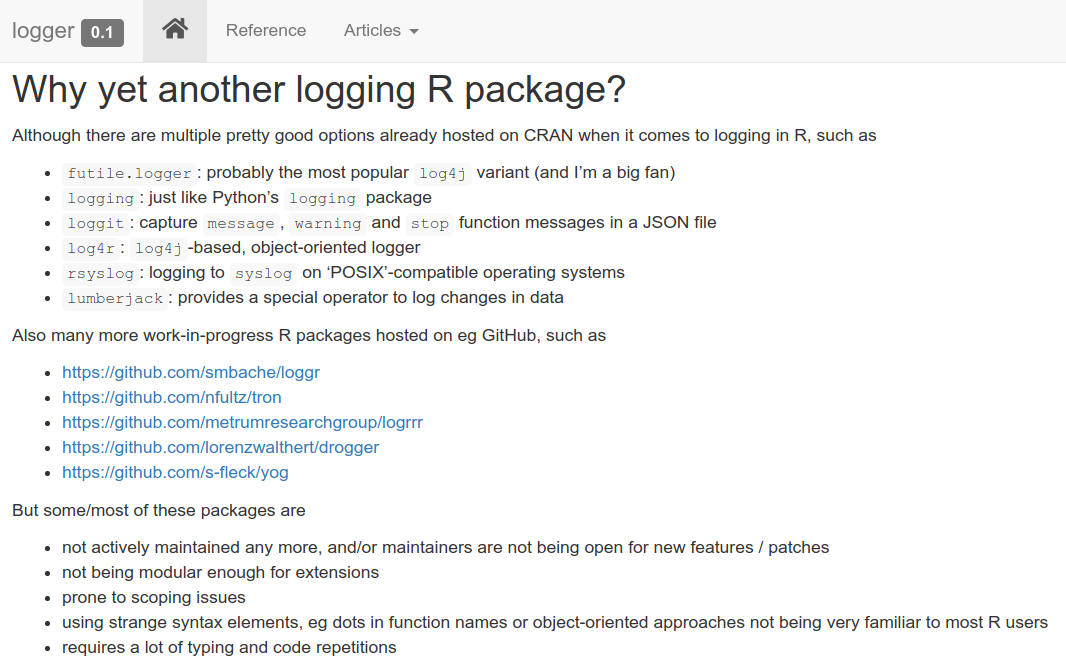
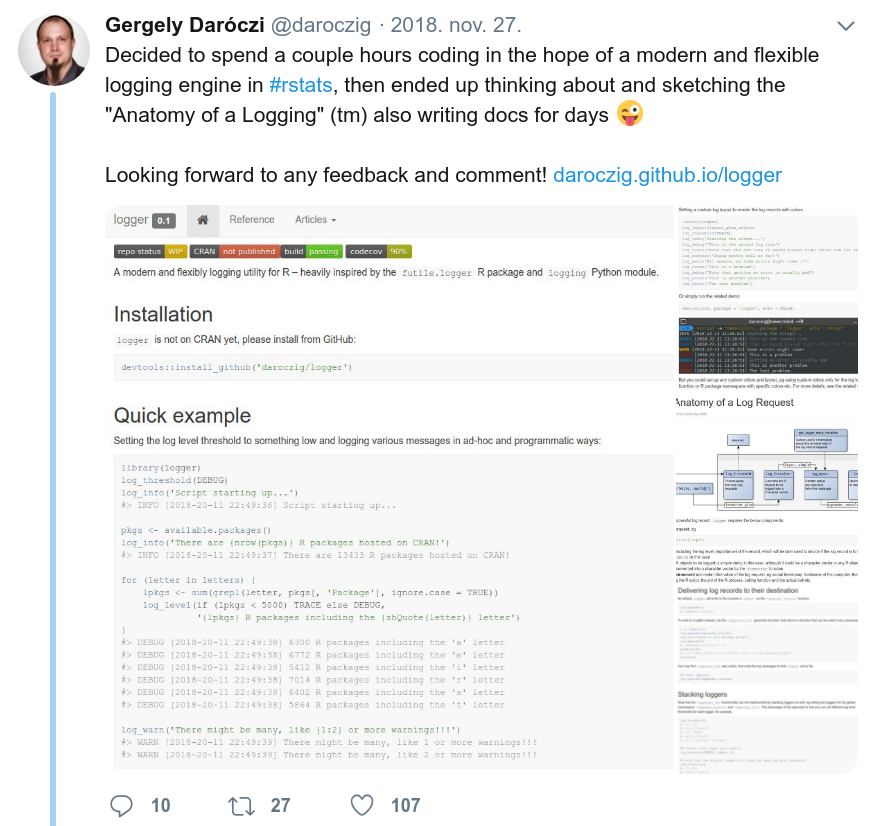
> demo(logger)
library(logger)
log_threshold(DEBUG)
log_info('Script starting up...')
#> INFO [2018-20-11 22:49:36] Script starting up...
pkgs <- available.packages()
log_info('There are {nrow(pkgs)} R packages hosted on CRAN!')
#> INFO [2018-20-11 22:49:37] There are 13433 R packages hosted on CRAN!
for (letter in letters) {
lpkgs <- sum(grepl(letter, pkgs[, 'Package'], ignore.case = TRUE))
log_level(if (lpkgs < 5000) TRACE else DEBUG,
'{lpkgs} R packages including the {shQuote(letter)} letter')
}
#> DEBUG [2018-20-11 22:49:38] 6300 R packages including the 'a' letter
#> DEBUG [2018-20-11 22:49:38] 6772 R packages including the 'e' letter
#> DEBUG [2018-20-11 22:49:38] 5412 R packages including the 'i' letter
#> DEBUG [2018-20-11 22:49:38] 7014 R packages including the 'r' letter
#> DEBUG [2018-20-11 22:49:38] 6402 R packages including the 's' letter
#> DEBUG [2018-20-11 22:49:38] 5864 R packages including the 't' letter> str(logger)
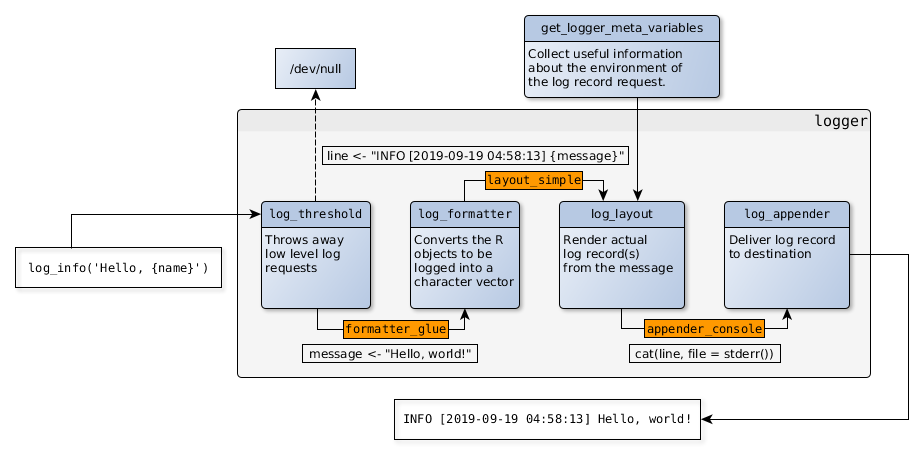
> library(logger)
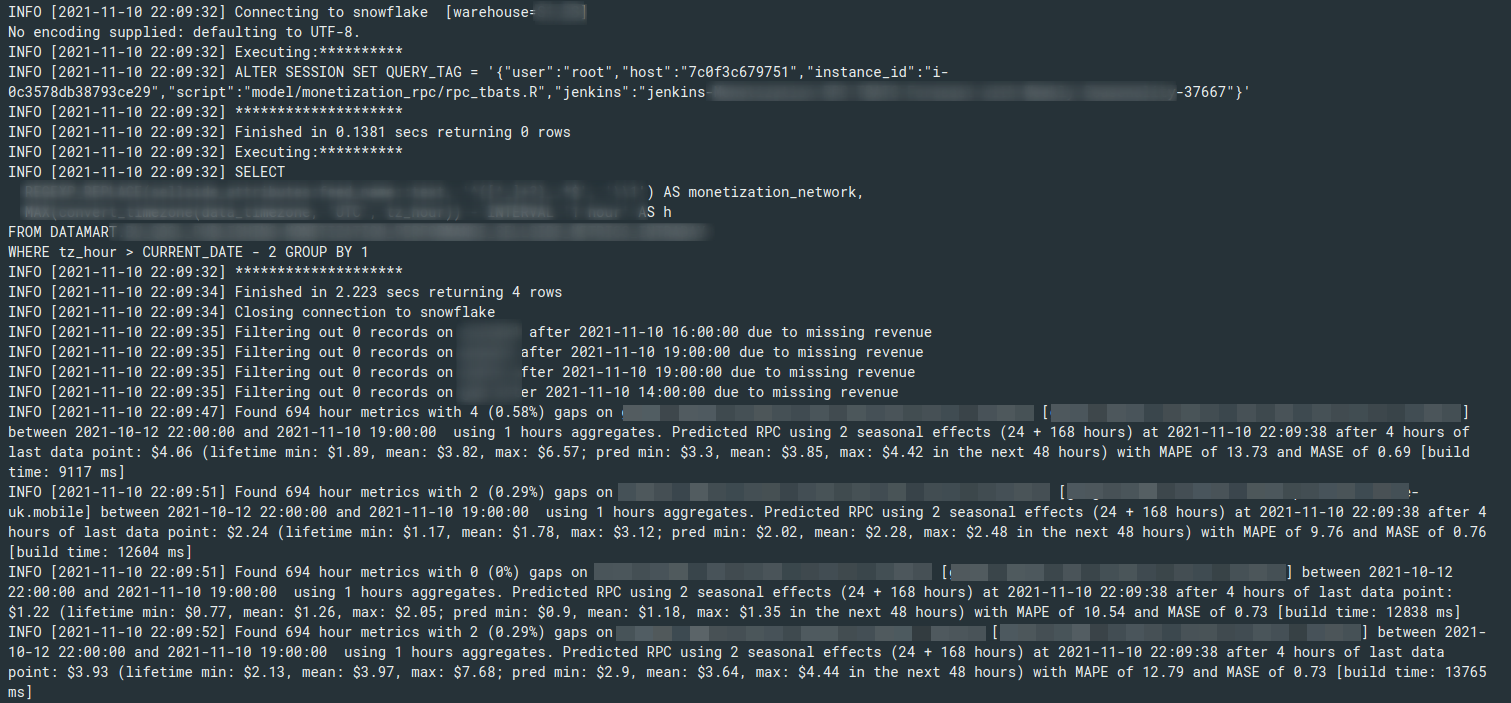
> library(logger)
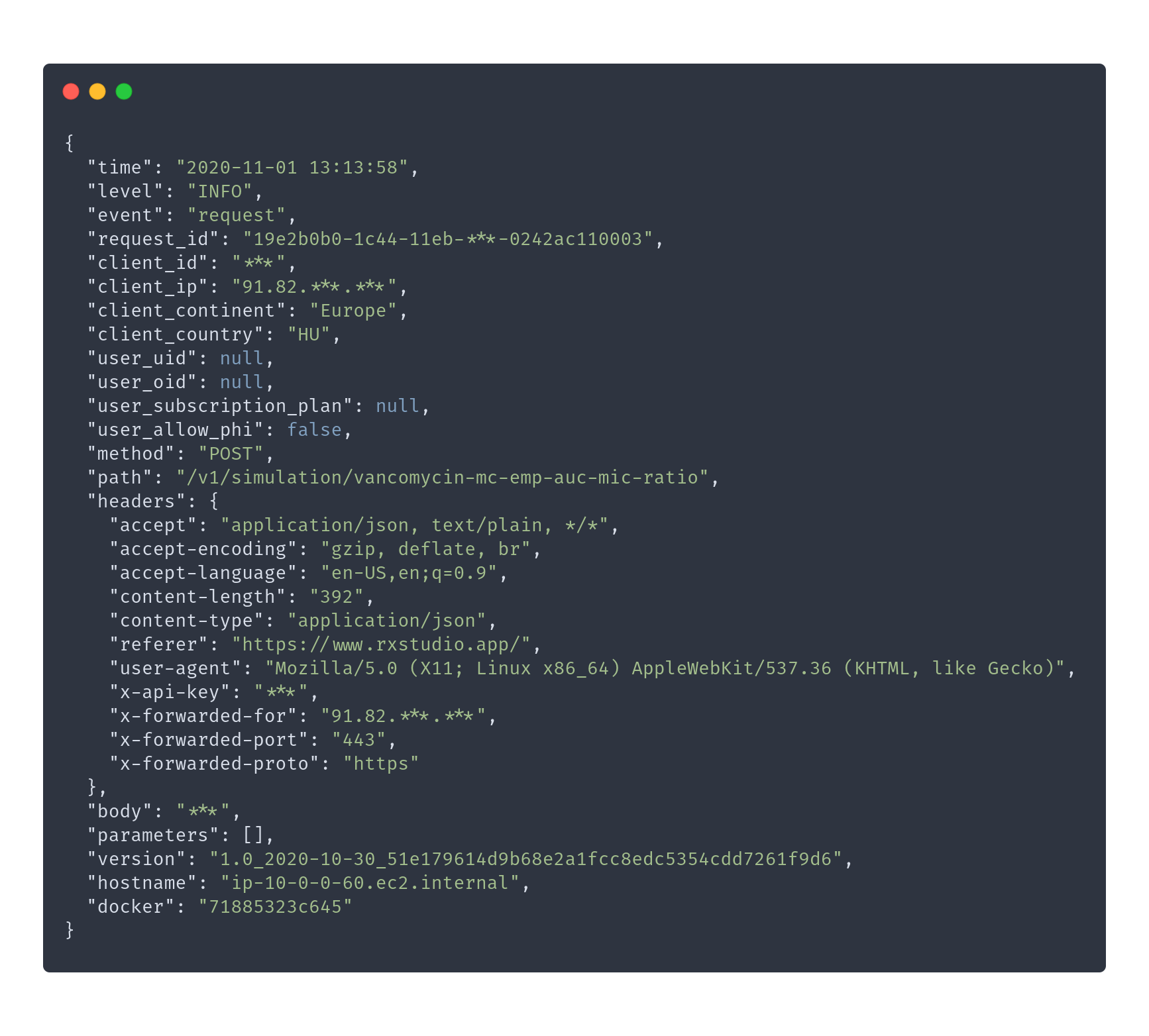
> library(logger)

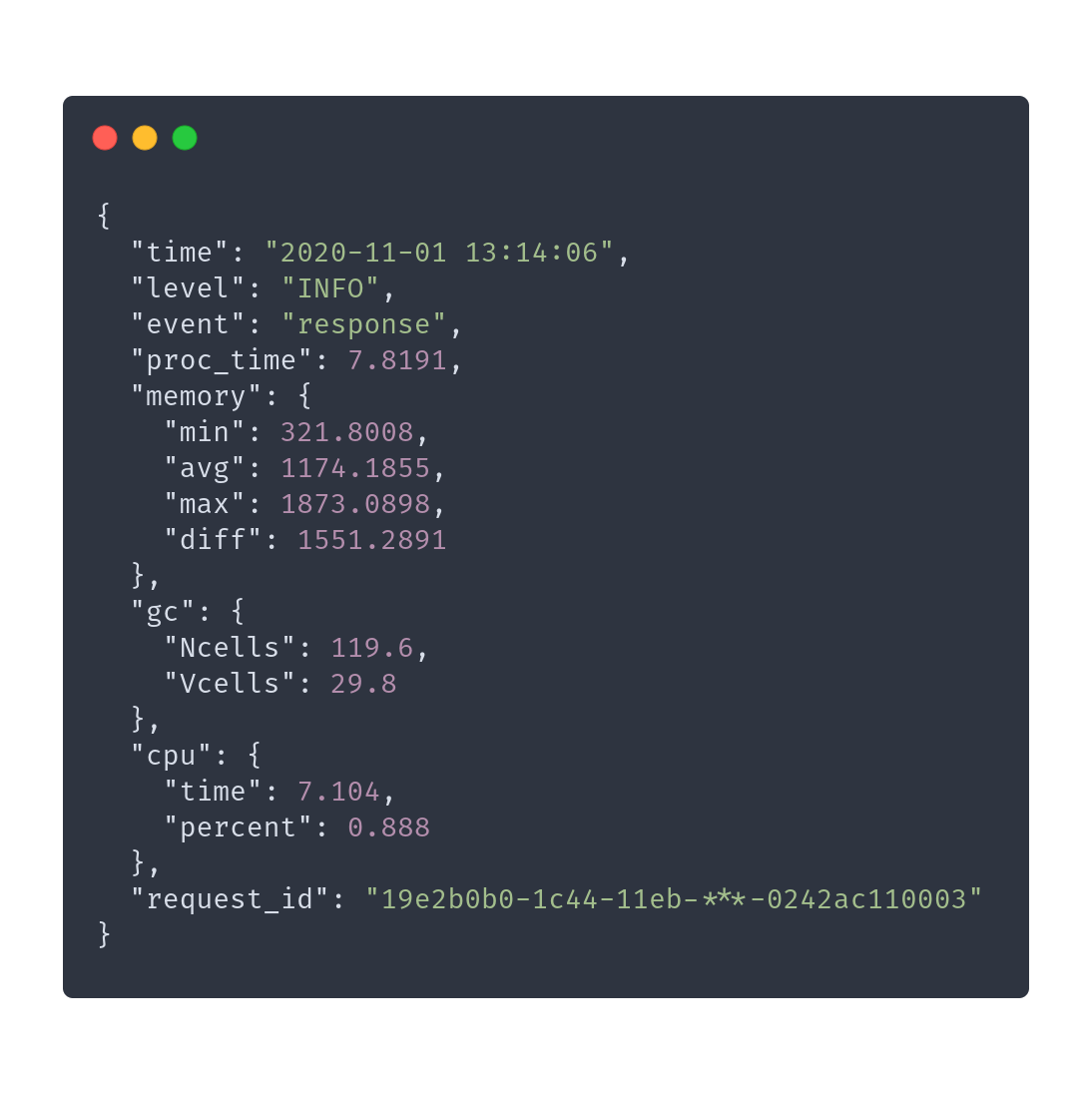
> requireNamespace(logger)
library(botor)
my_mtcars <- s3_read('s3://botor/example-data/mtcars.csv', read.csv)
#> DEBUG [2019-09-19 04:46:57] Downloaded 1303 bytes from s3://botor/example-data/mtcars.csv
#> and saved at '/tmp/RtmpLW4bY4/file63ff42ed2fe1'log_threshold(TRACE, namespace = 'botor')
my_mtcars <- s3_read('s3://botor/example-data/mtcars.csv.gz',
read.csv, extract = 'gzip')
#> TRACE [2019-09-19 04:48:02] Downloading s3://botor/example-data/mtcars.csv.gz to
#> '/tmp/RtmpLW4bY4/file63ff17e137e9' ...
#> DEBUG [2019-09-19 04:48:03] Downloaded 567 bytes from s3://botor/example-data/mtcars.csv.gz
#> and saved at '/tmp/RtmpLW4bY4/file63ff17e137e9'
#> TRACE [2019-09-19 04:48:03] Decompressed /tmp/RtmpLW4bY4/file63ff17e137e9 via gzip
#> from 567 to 1303 bytes
#> TRACE [2019-09-19 04:48:03] Deleted /tmp/RtmpLW4bY4/file63ff17e137e9> requireNamespace(logger)
library(dbr)
str(db_query('SELECT 42', 'sqlite'))
#> INFO [2018-07-11 17:07:12] Connecting to sqlite
#> INFO [2018-07-11 17:07:12] Executing:**********
#> INFO [2018-07-11 17:07:12] SELECT 42
#> INFO [2018-07-11 17:07:12] ********************
#> INFO [2018-07-11 17:07:12] Finished in 0.0007429 secs returning 1 rows
#> INFO [2018-07-11 17:07:12] Closing connection to sqlite
#> 'data.frame': 1 obs. of 1 variable:
#> $ 42: int 42
#> - attr(*, "when")= POSIXct, format: "2018-07-11 17:07:12"
#> - attr(*, "db")= chr "sqlite"
#> - attr(*, "time_to_exec")=Class 'difftime' atomic [1:1] 0.000743
#> .. ..- attr(*, "units")= chr "secs"
#> - attr(*, "statement")= chr "SELECT 42"> log_shiny_input_changes()
library(shiny)
ui <- bootstrapPage(
numericInput('mean', 'mean', 0),
numericInput('sd', 'sd', 1),
textInput('title', 'title', 'title'),
plotOutput('plot')
)
server <- function(input, output) {
logger::log_shiny_input_changes(input)
output$plot <- renderPlot({
hist(rnorm(1e3, input$mean, input$sd), main = input$title)
})
}
shinyApp(ui = ui, server = server)> log_shiny_input_changes()
Listening on http://127.0.0.1:8080
INFO [2019-07-11 16:59:17] Default Shiny inputs initialized: {"mean":0,"title":"title","sd":1}
INFO [2019-07-11 16:59:26] Shiny input change detected on mean: 0 -> 1
INFO [2019-07-11 16:59:27] Shiny input change detected on mean: 1 -> 2
INFO [2019-07-11 16:59:27] Shiny input change detected on mean: 2 -> 3
INFO [2019-07-11 16:59:27] Shiny input change detected on mean: 3 -> 4
INFO [2019-07-11 16:59:27] Shiny input change detected on mean: 4 -> 5
INFO [2019-07-11 16:59:27] Shiny input change detected on mean: 5 -> 6
INFO [2019-07-11 16:59:27] Shiny input change detected on mean: 6 -> 7
INFO [2019-07-11 16:59:29] Shiny input change detected on sd: 1 -> 2
INFO [2019-07-11 16:59:29] Shiny input change detected on sd: 2 -> 3
INFO [2019-07-11 16:59:29] Shiny input change detected on sd: 3 -> 4
INFO [2019-07-11 16:59:29] Shiny input change detected on sd: 4 -> 5
INFO [2019-07-11 16:59:29] Shiny input change detected on sd: 5 -> 6
INFO [2019-07-11 16:59:29] Shiny input change detected on sd: 6 -> 7
INFO [2019-07-11 16:59:34] Shiny input change detected on title: title -> sfdsadsads> log_appender(appender_async(…))
- create a local, disk-based storage for the message queue via
txtq - start a background process for the async execution of the message queue with
callr - loads minimum required packages in the background process
- connects to the message queue from the background process
- pass actual
appenderfunction to the background process (serialized to disk) - pass parameters of the async appender to the background process (eg batch size)
- start infinite loop processing log records
- check if background process still works …
> demo(logger)
- log levels
- log message formatter functions
- log record layout functions
- log message delivery functions
- namespaces
- stacking
- helpers
- …
Check out the “Getting things logged” talk at RStudio::conf(2020)!
> sample(projects, size = 5)
> sample(projects, size = 5)
> sample(projects, size = 5)
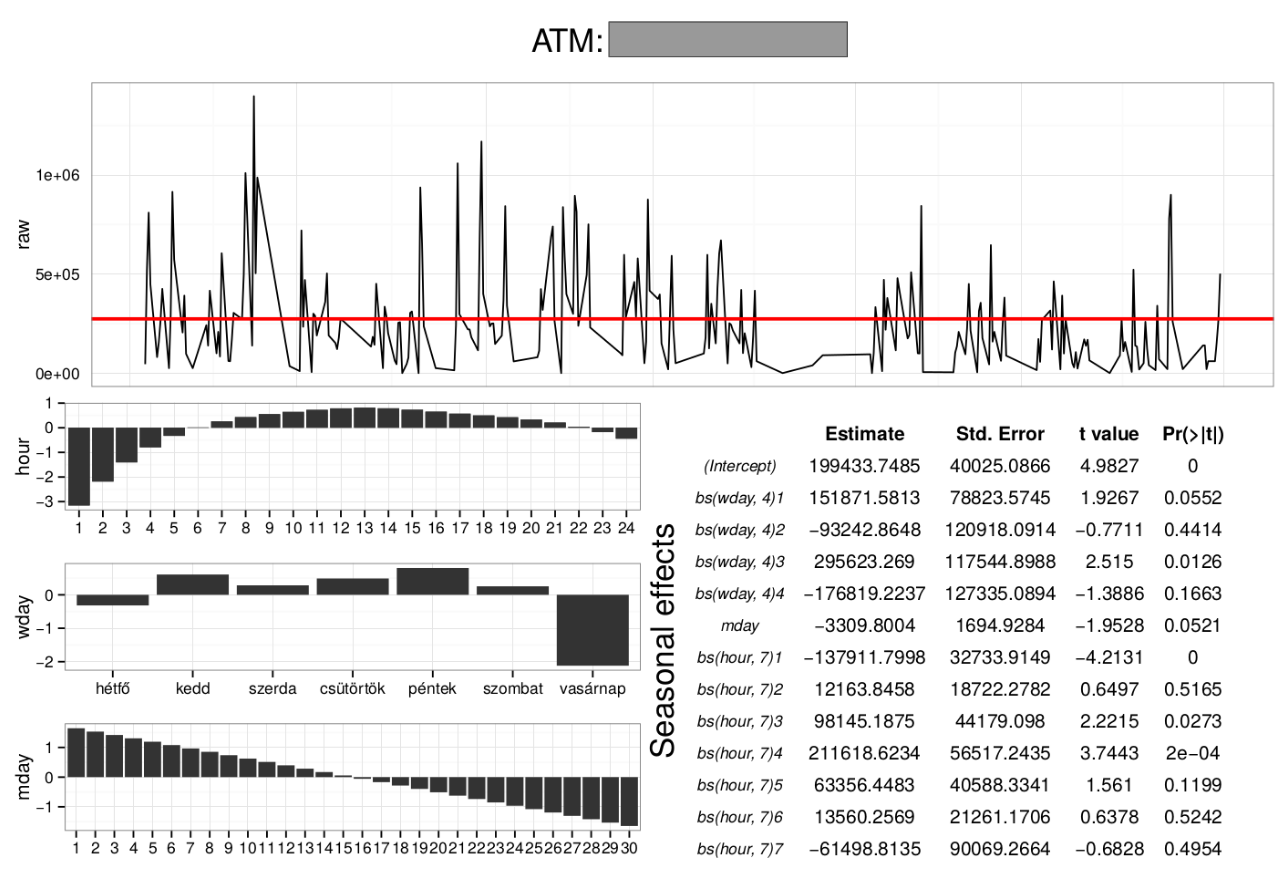
> sample(projects, size = 5)
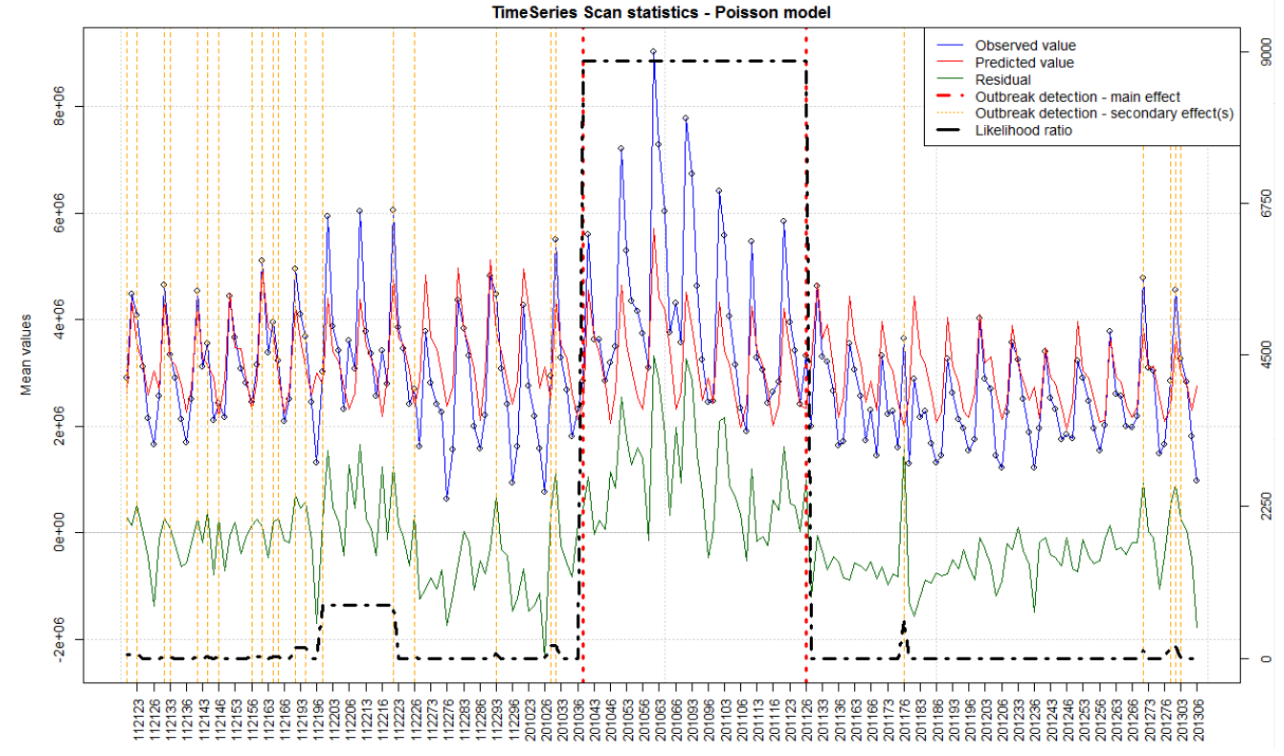
> sample(projects, size = 5)
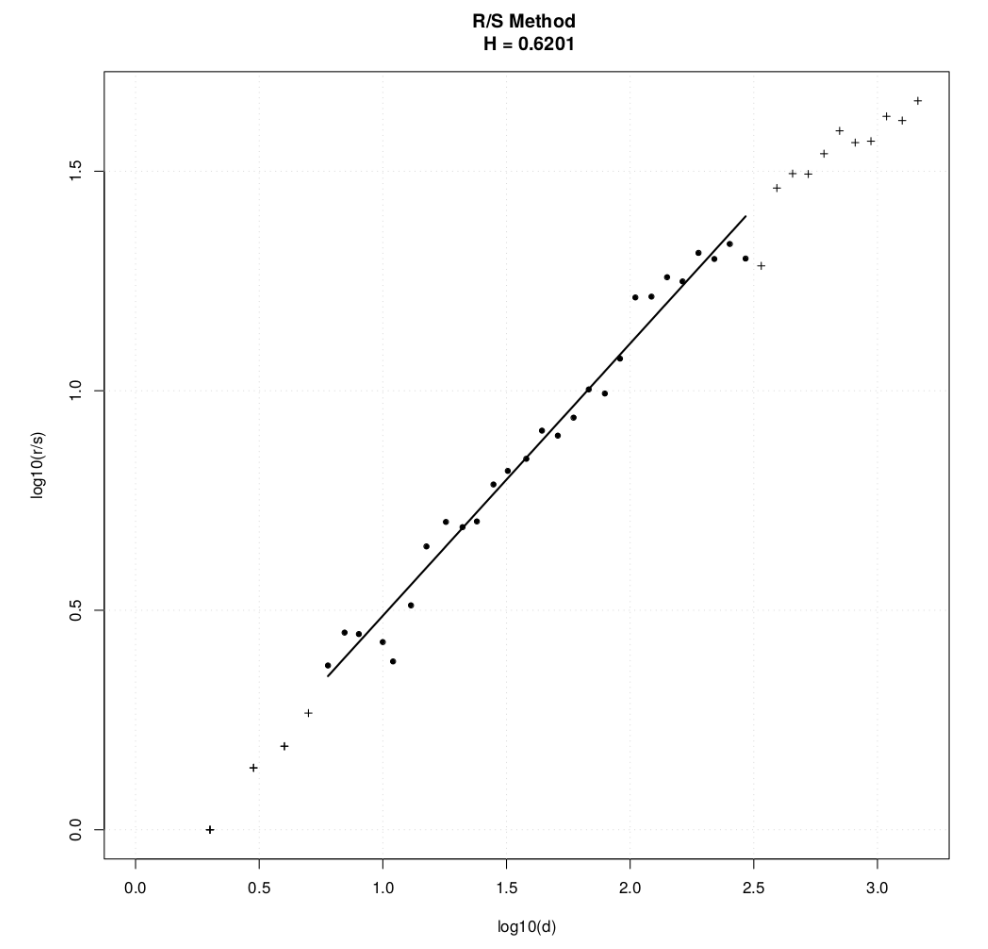
> sample(projects, size = 5)

> sample(projects, size = 5)

> sample(projects, size = 5)

> usethis::create_startup(…)

> demo(“rapporter.net”)
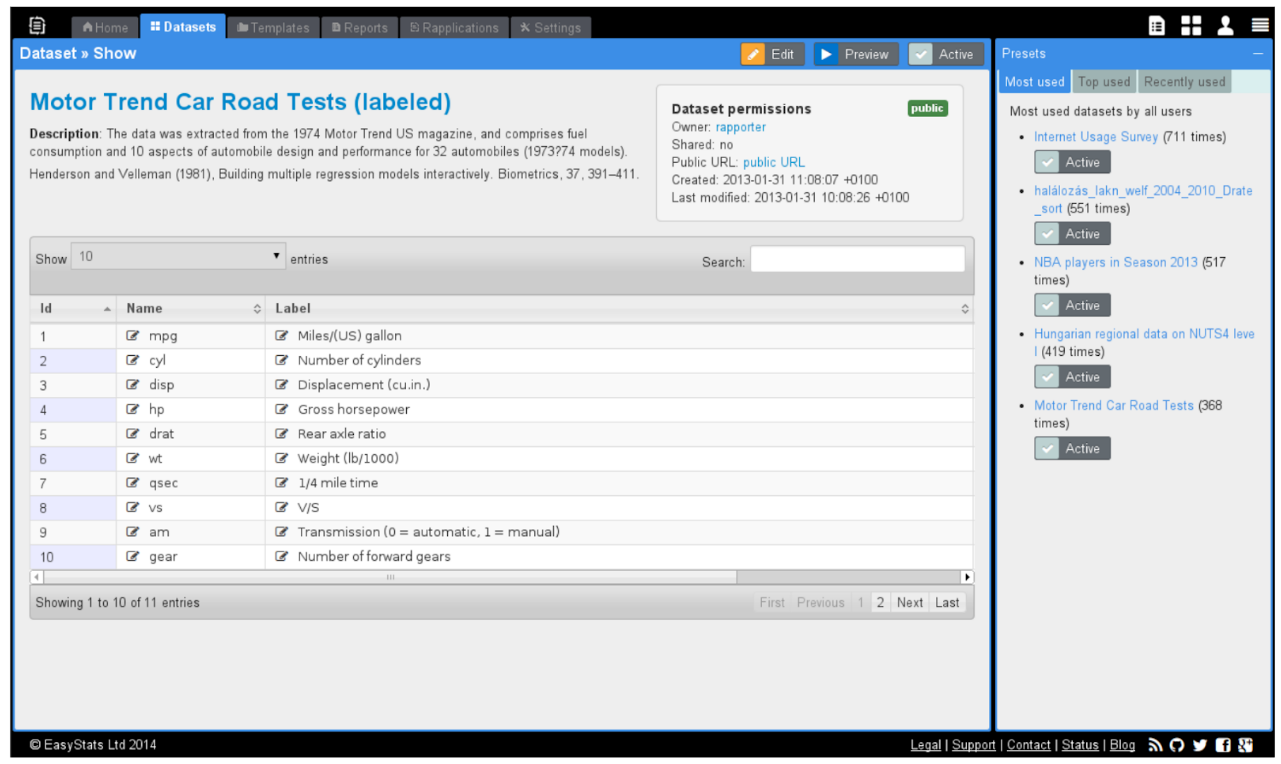
> demo(“rapporter.net”)
> demo(“rapporter.net”)
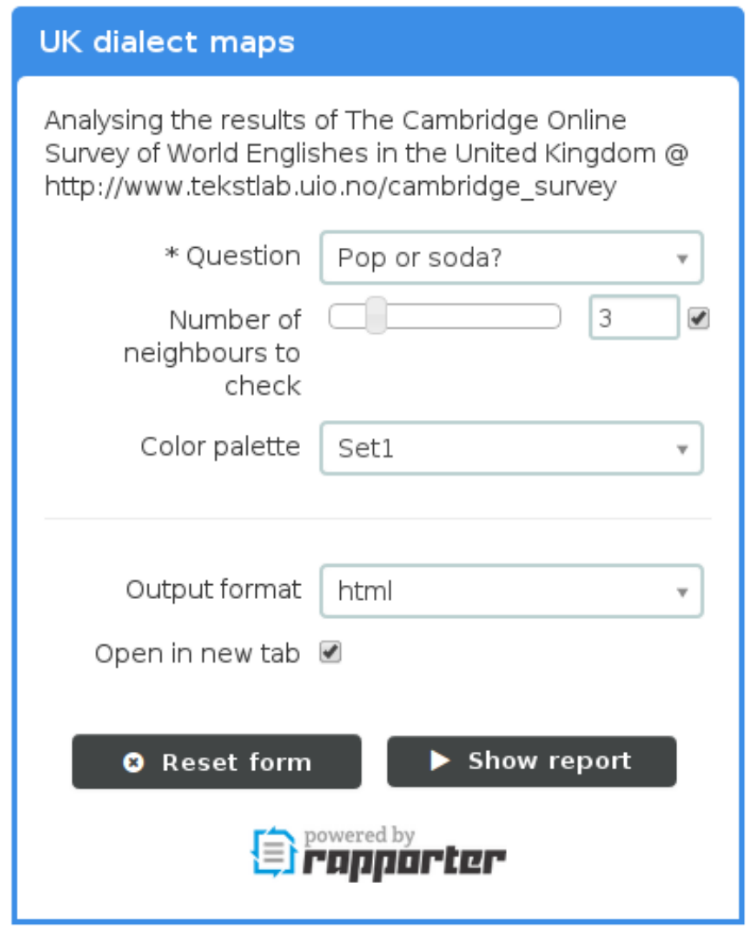
> demo(“rapporter.net”)
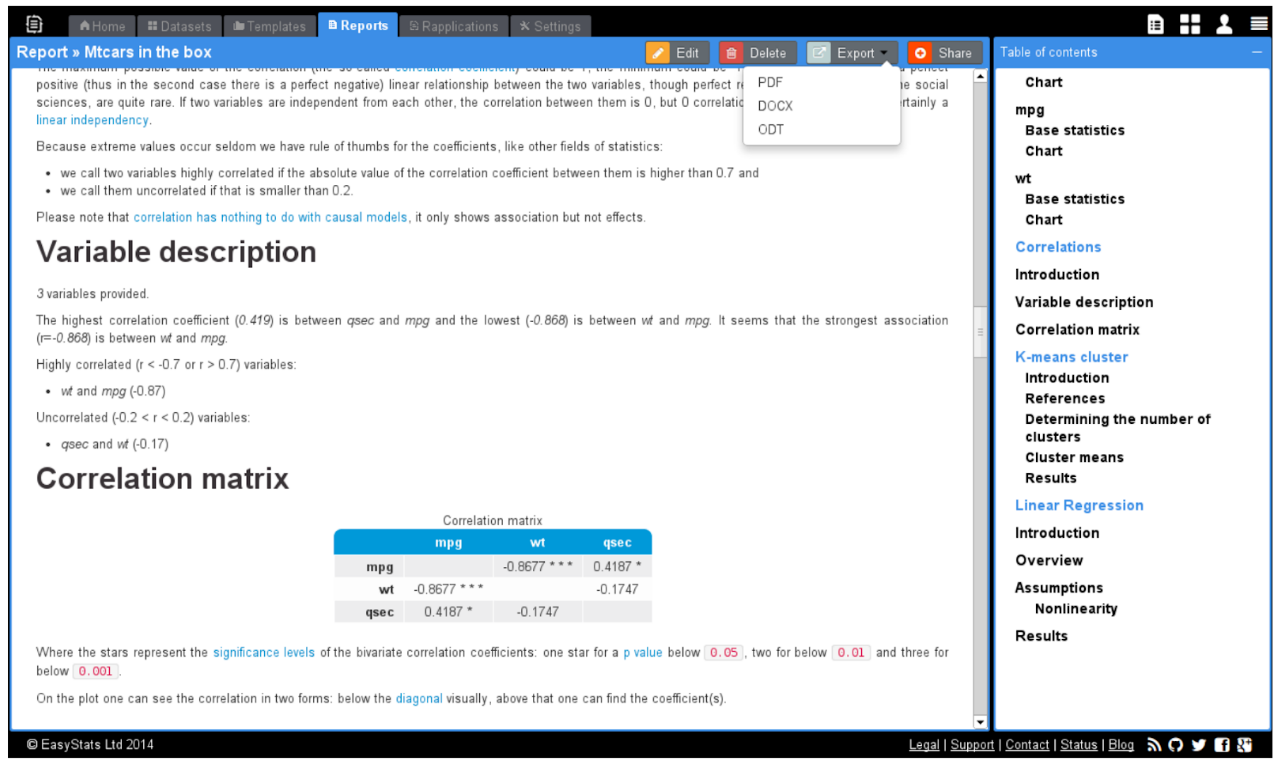
> demo(“rapporter.net”)
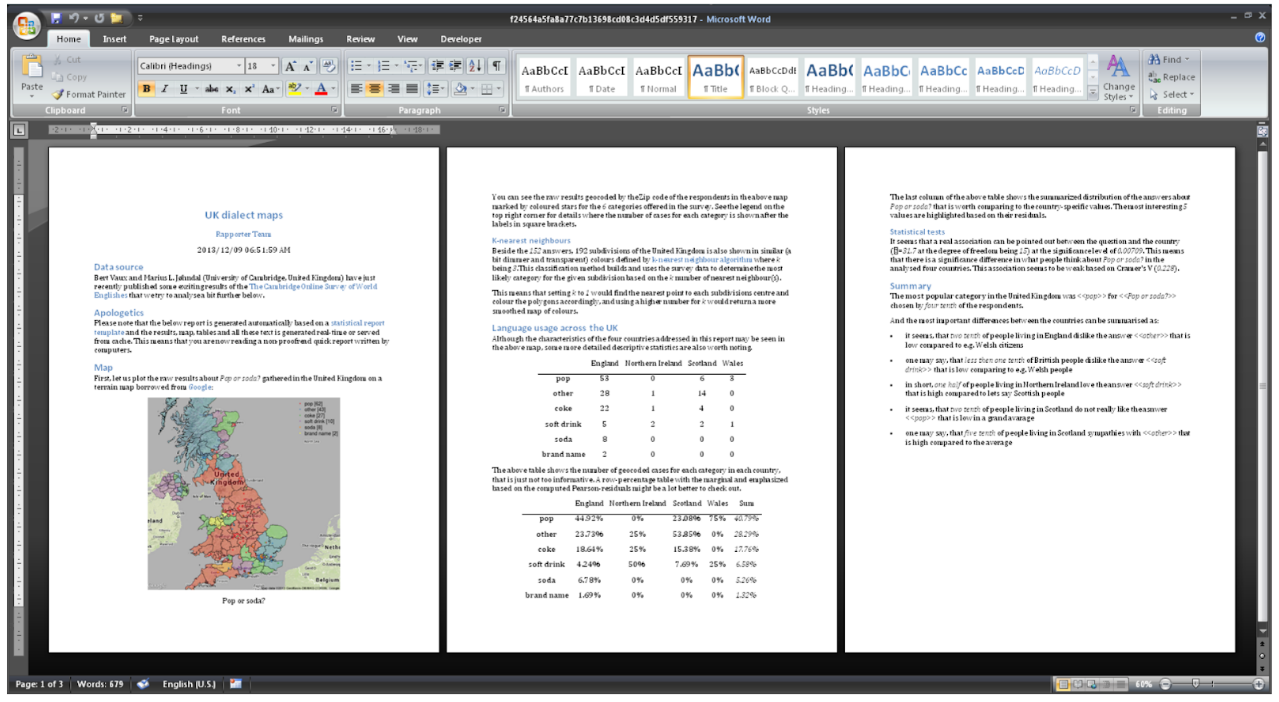
> demo(“rapporter.net”)

> demo(“rapporter.net”)

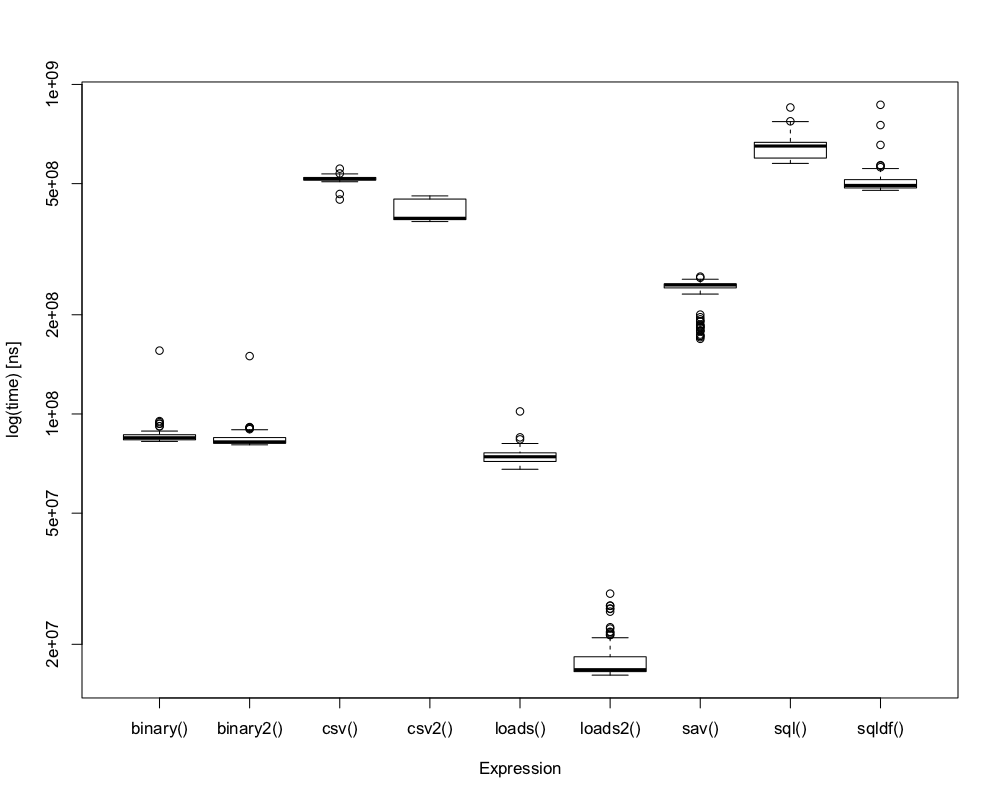
> microbenchmark::microbenchmark(…)

> microbenchmark::microbenchmark(…)
## "benchmark" done with: a list holding 1500 elements (all holding 4 chars)
> system.time(rjson::toJSON(rjson::fromJSON(x)))
user system elapsed
10.000 0.010 10.073
> system.time(RJSONIO::toJSON(RJSONIO::fromJSON(x)))
user system elapsed
0.797 0.003 0.812
## RJSONIO all the way, but it is still slow :(> microbenchmark::microbenchmark(…)
> txt <- paste(sample(letters, 1e3, replace = TRUE), collapse = '')
> file <- tempfile()
> cat(txt, file = file)
> library(microbenchmark)
> ca <- function() caTools:::base64encode(txt)
> base64 <- function(letters) base64::encode(file)
> curl <- function() RCurl:::base64Encode(txt)
> microbenchmark(ca(), base64(), curl(), times = 1e3)
Unit: microseconds
expr min lq mean median uq max neval cld
ca() 261.102 277.7570 338.45346 290.5145 318.3690 2485.63 1000 b
base64() 308.341 328.0395 392.70745 342.2250 380.6865 4295.81 1000 c
curl() 73.837 79.4285 92.78261 84.8370 90.9490 1226.68 1000 a> microbenchmark::microbenchmark(…)
> microbenchmark::microbenchmark(…)
> help(microbenchmark)

> microbenchmark::microbenchmark(…)
> microbenchmark::microbenchmark(…)
> plot(microbenchmark(…))
> ?alias

Source: Junior dev being awesome
> Sys.setlocale(‘en-US’)
> order(‘I-heart-R’)

> order(sample(n = 3))

> str(platform)
> str(stack)

> debugonce()
> usethis::create_package(‘fbRads’)

> usethis::create_packages(‘AWR’)

> predict(gbm, newdata = tx)
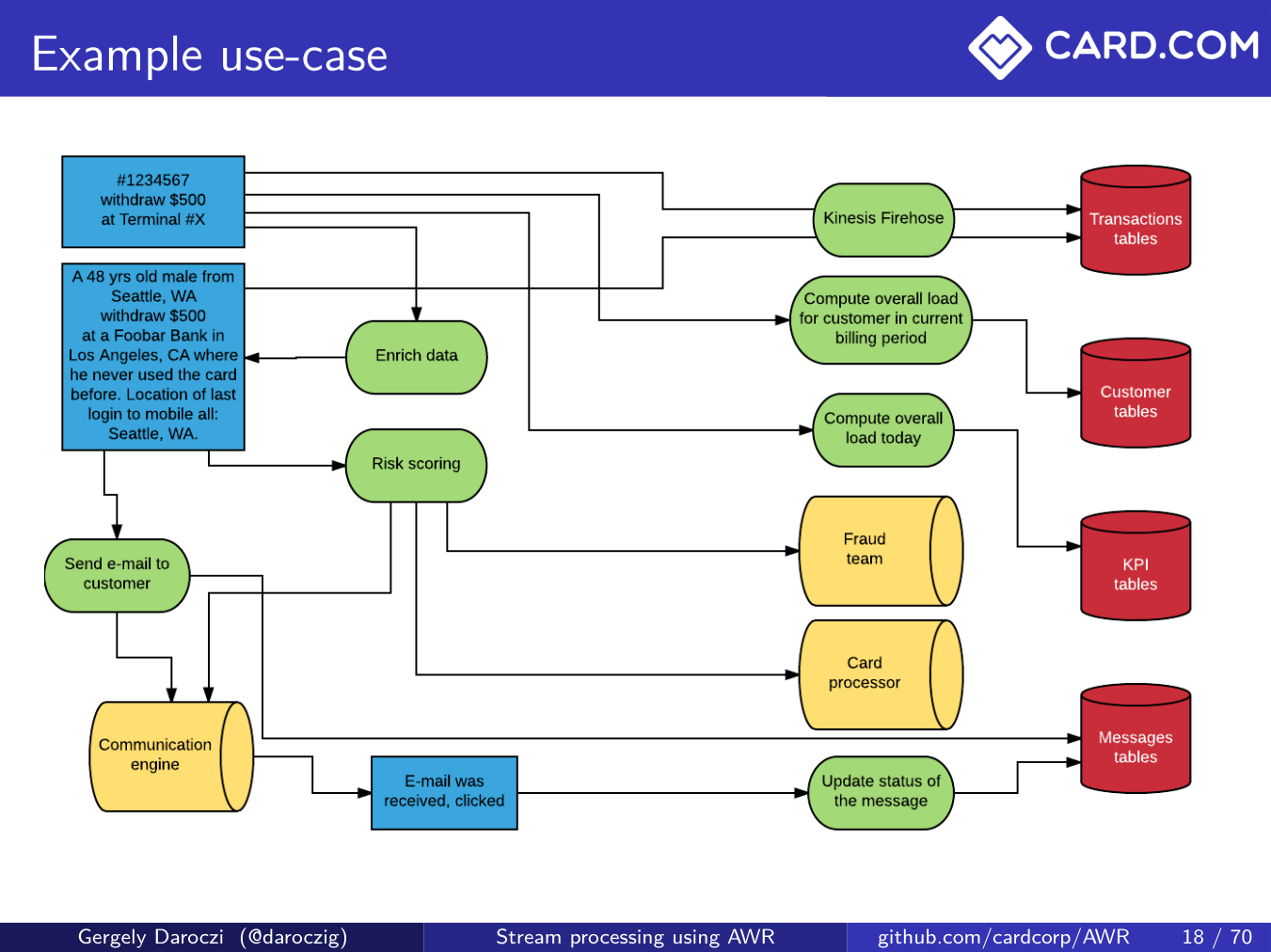
> git2r::commits()
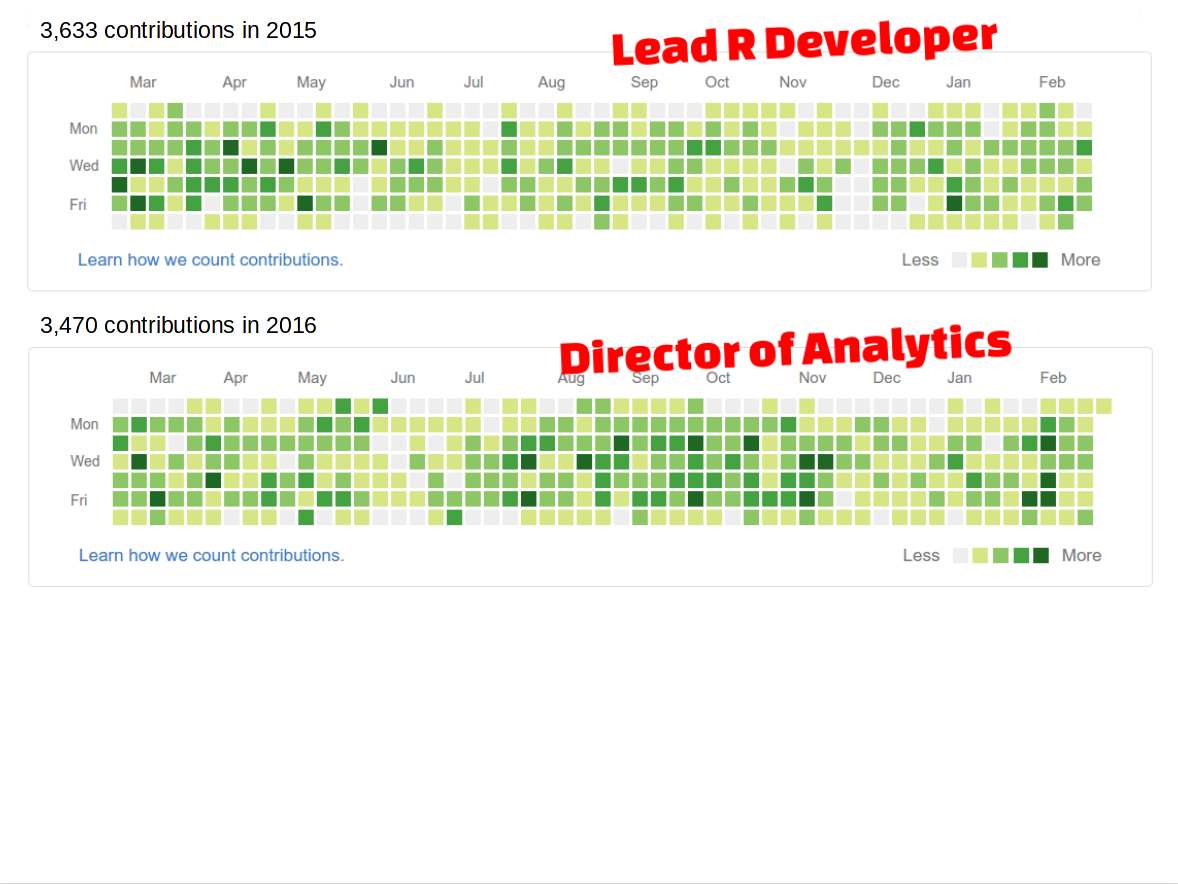
> git2r::commits()
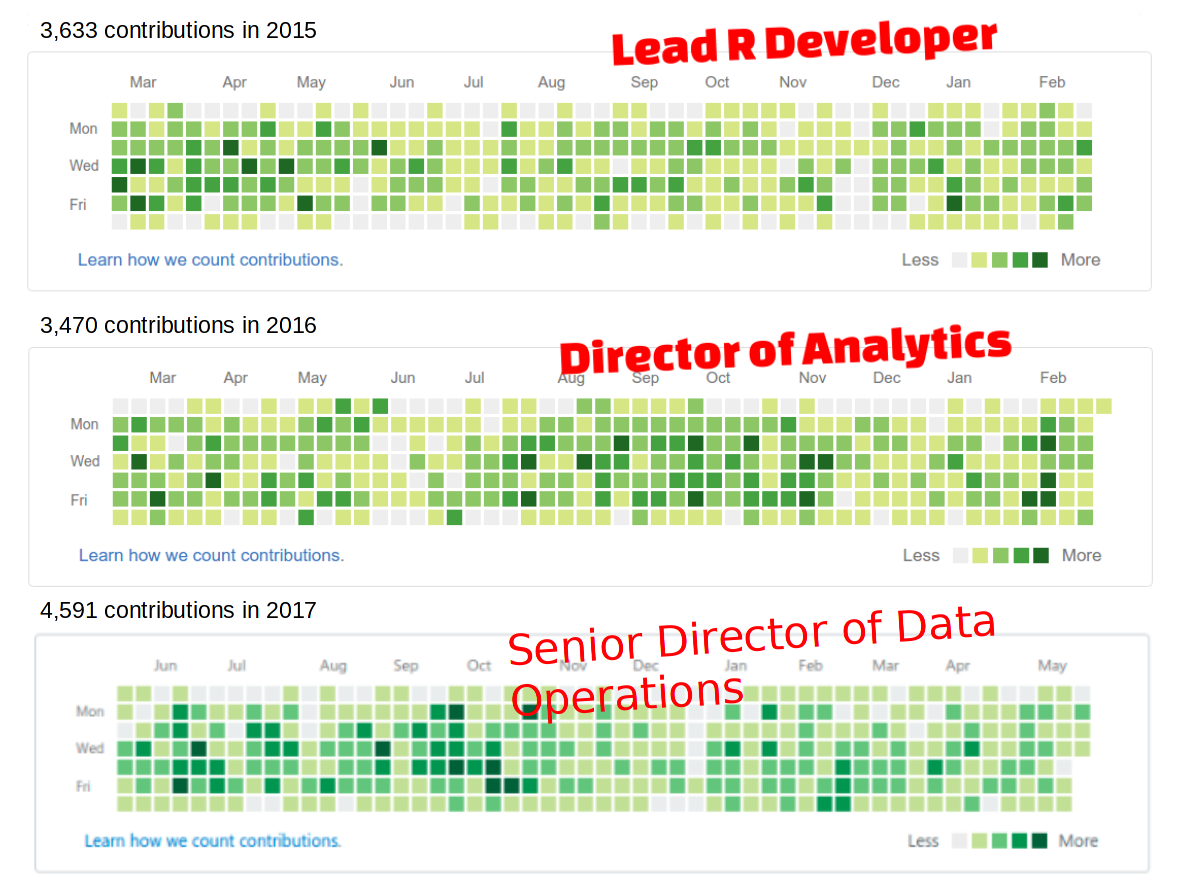
$ groups
$ groups
$ research()
> licence()
> licence()
> library(AWR.Snowflake)
> microbenchmark()
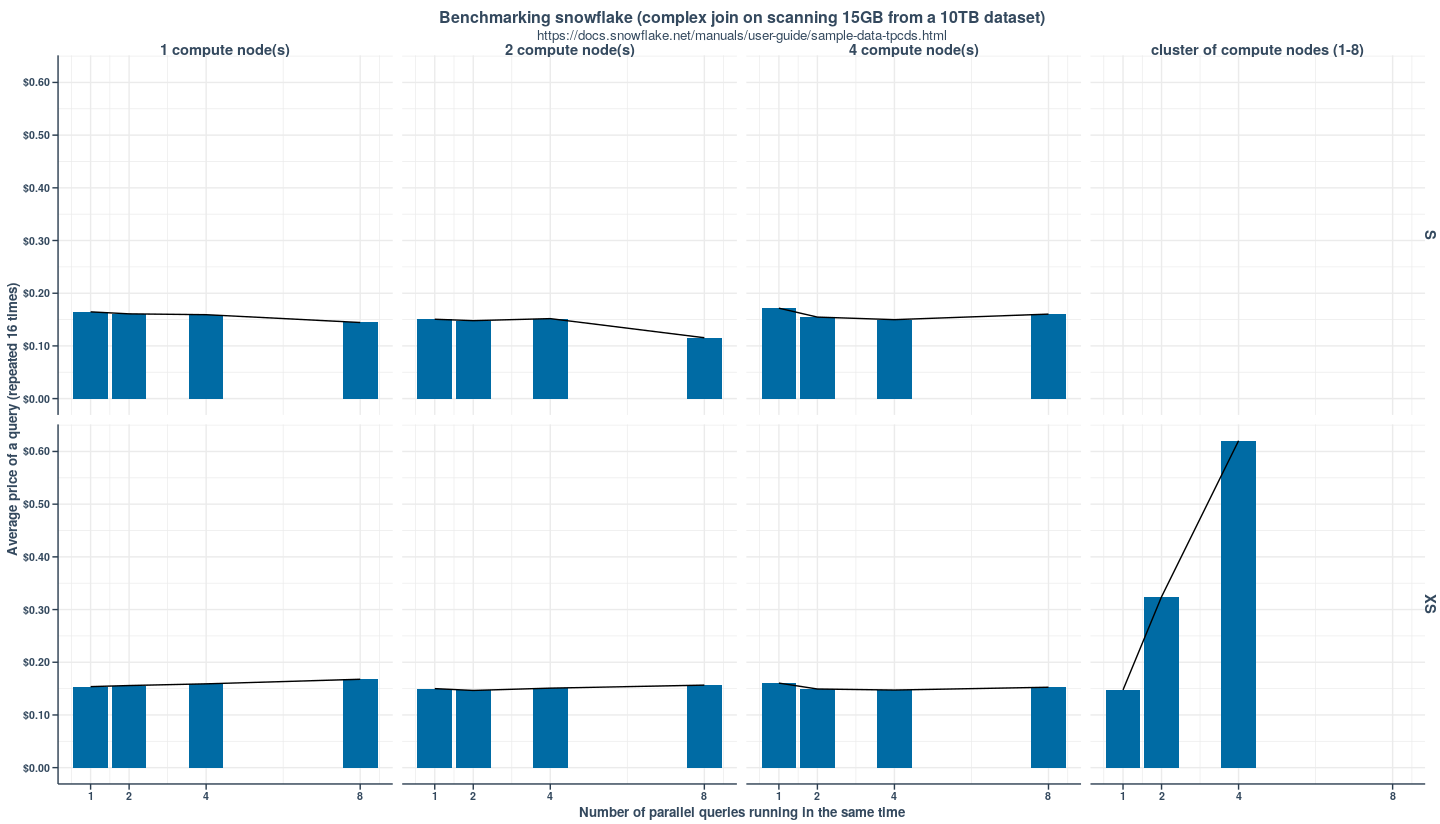
> options(error = browser())
> compare(‘spark’, ‘K8s’, …)
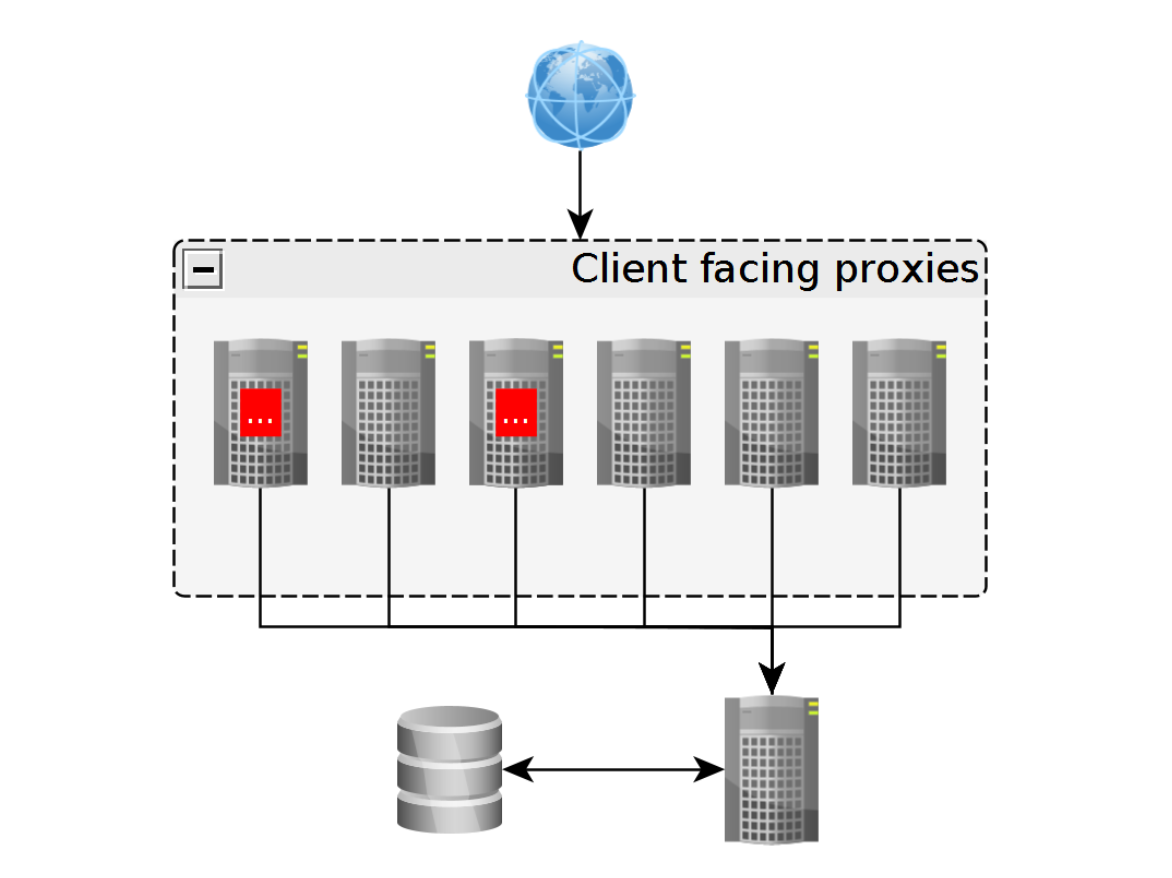
> compare(‘spark’, ‘K8s’, …)
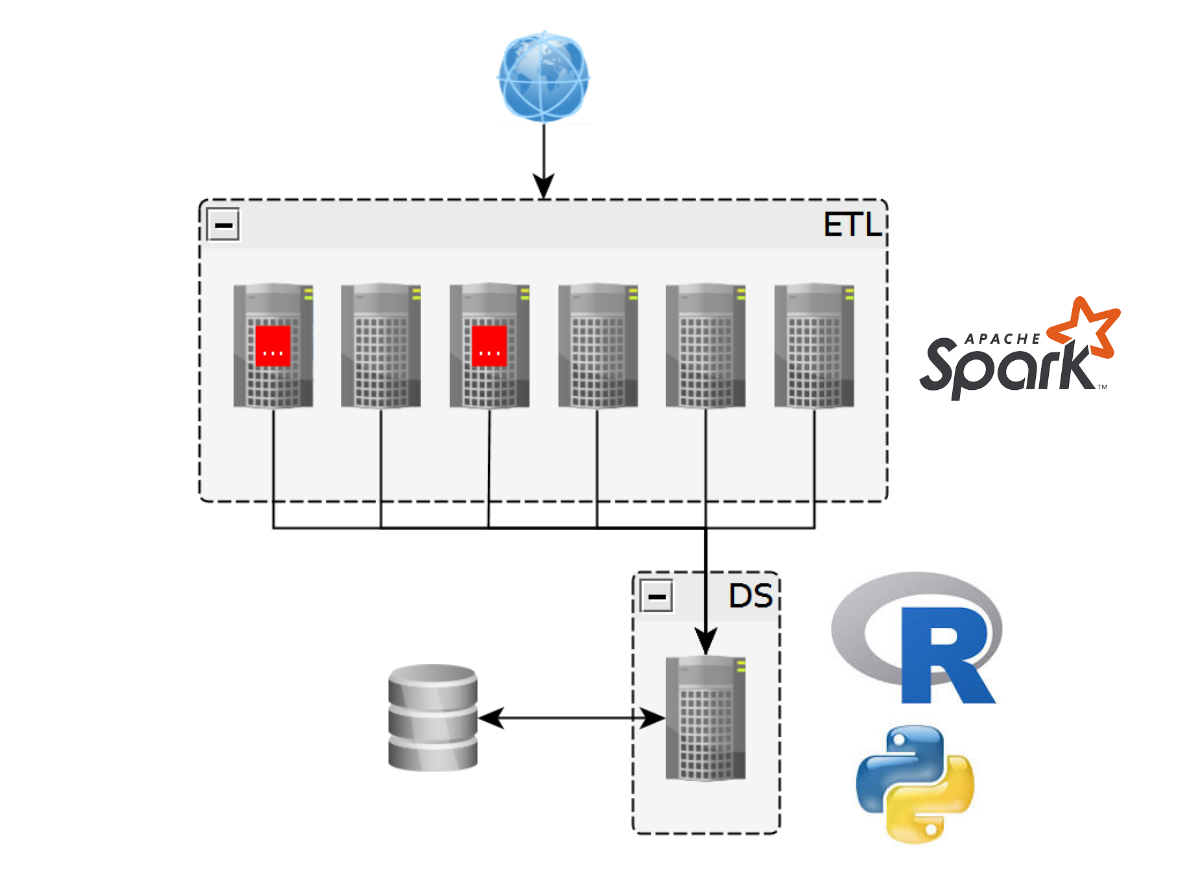
> ls(envir = ‘jobs’)
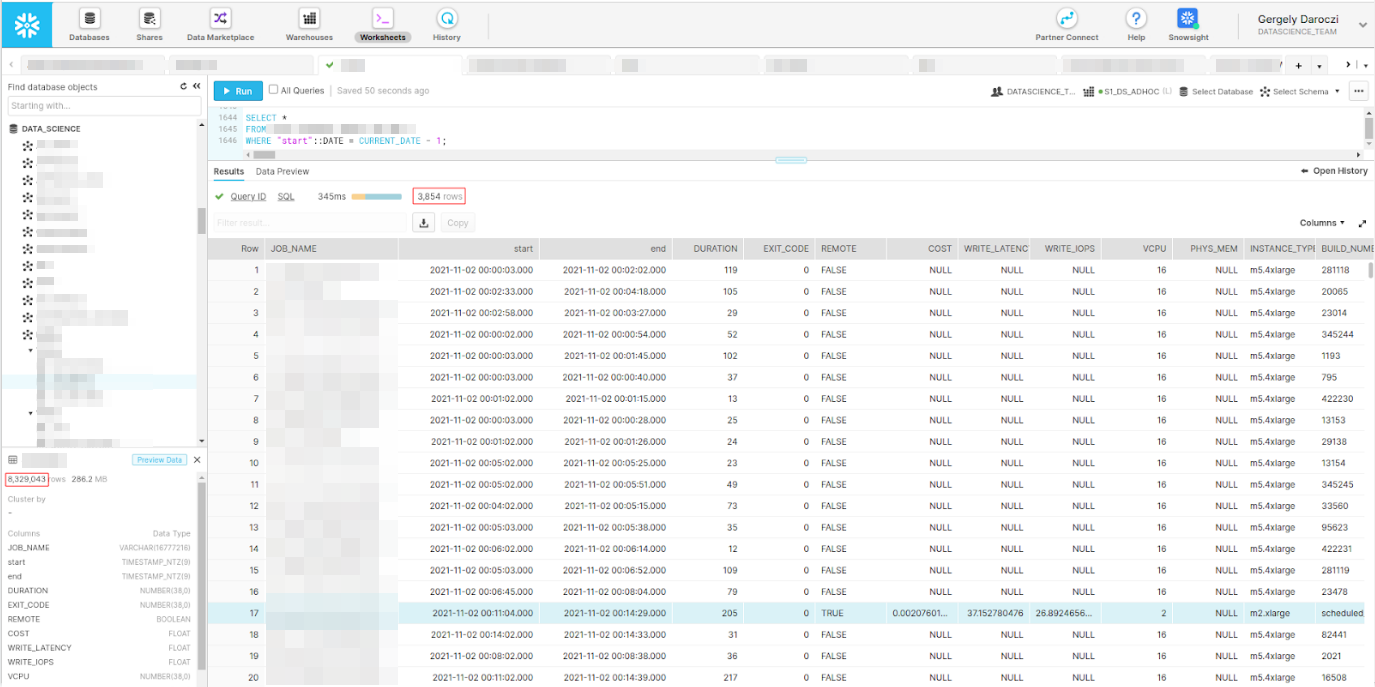
> get(‘job’)
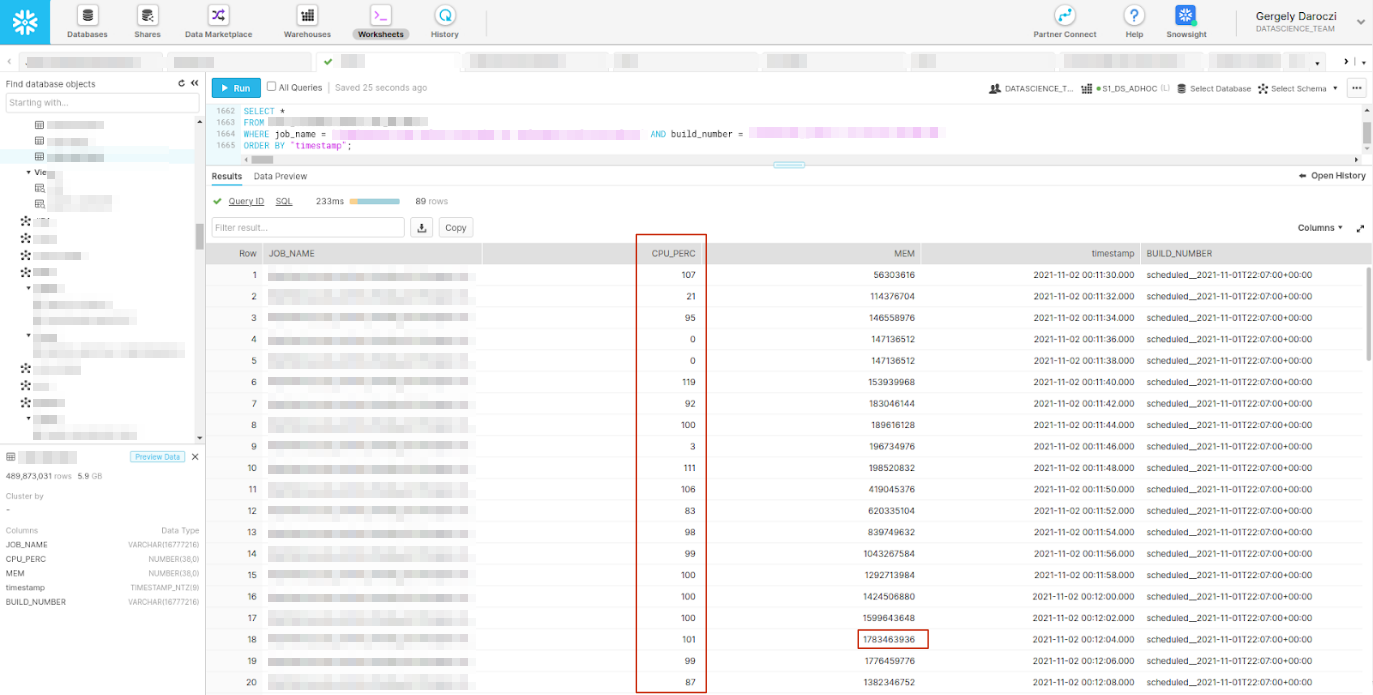
> eval()
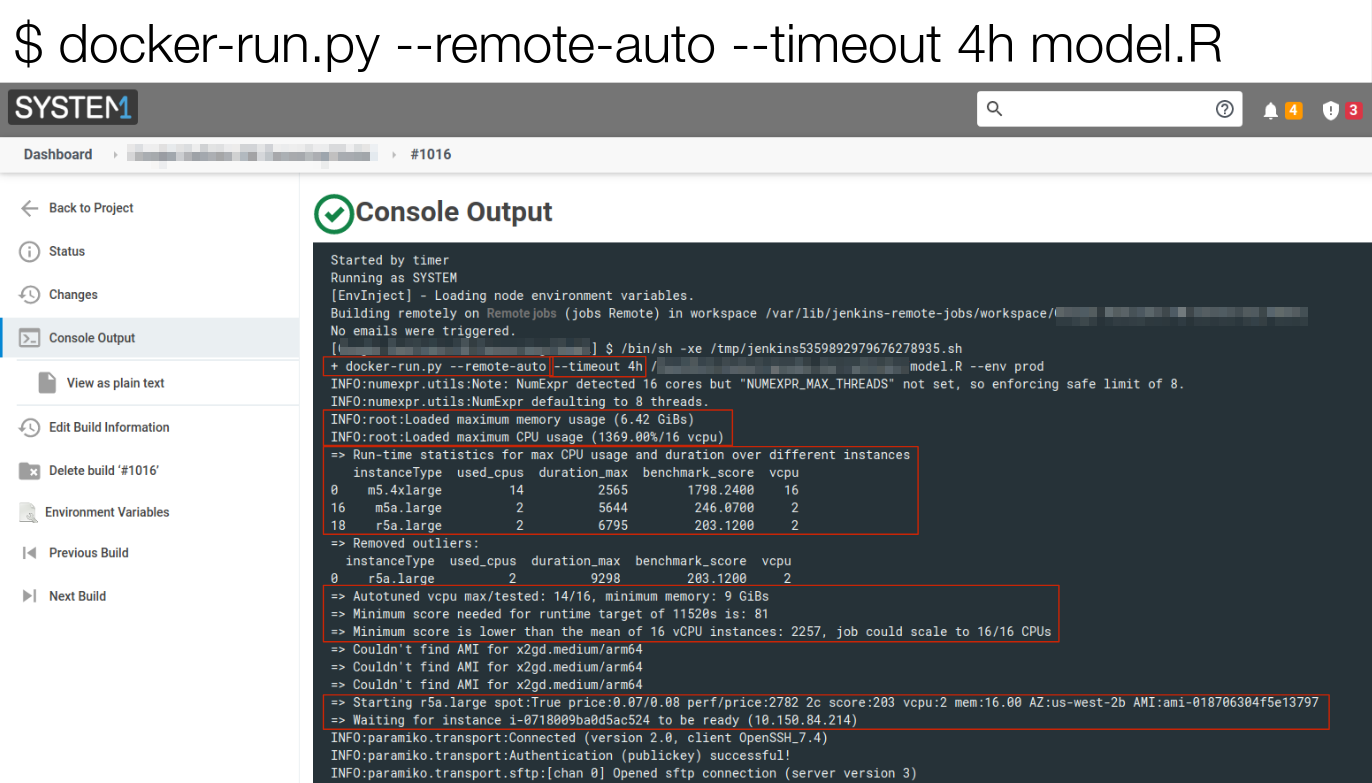
> eval()
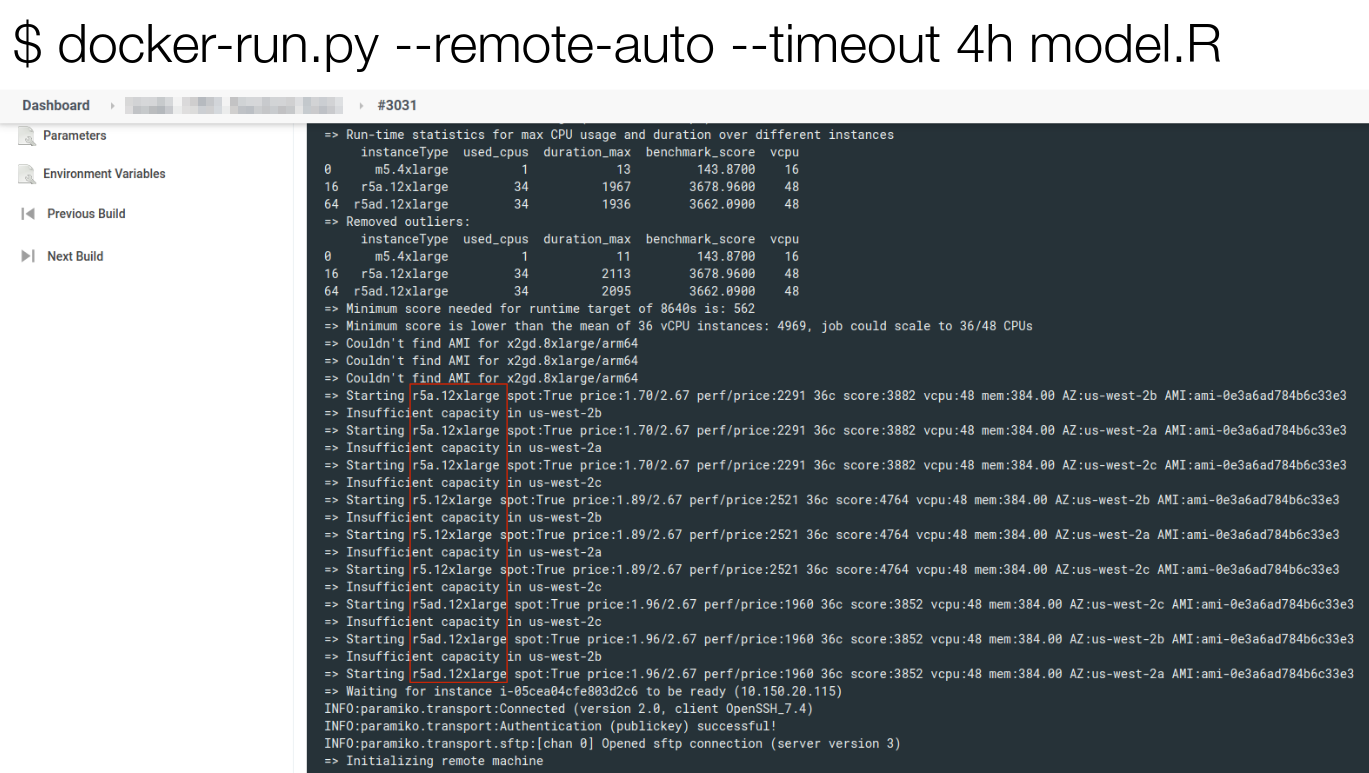
> source()

> licence()
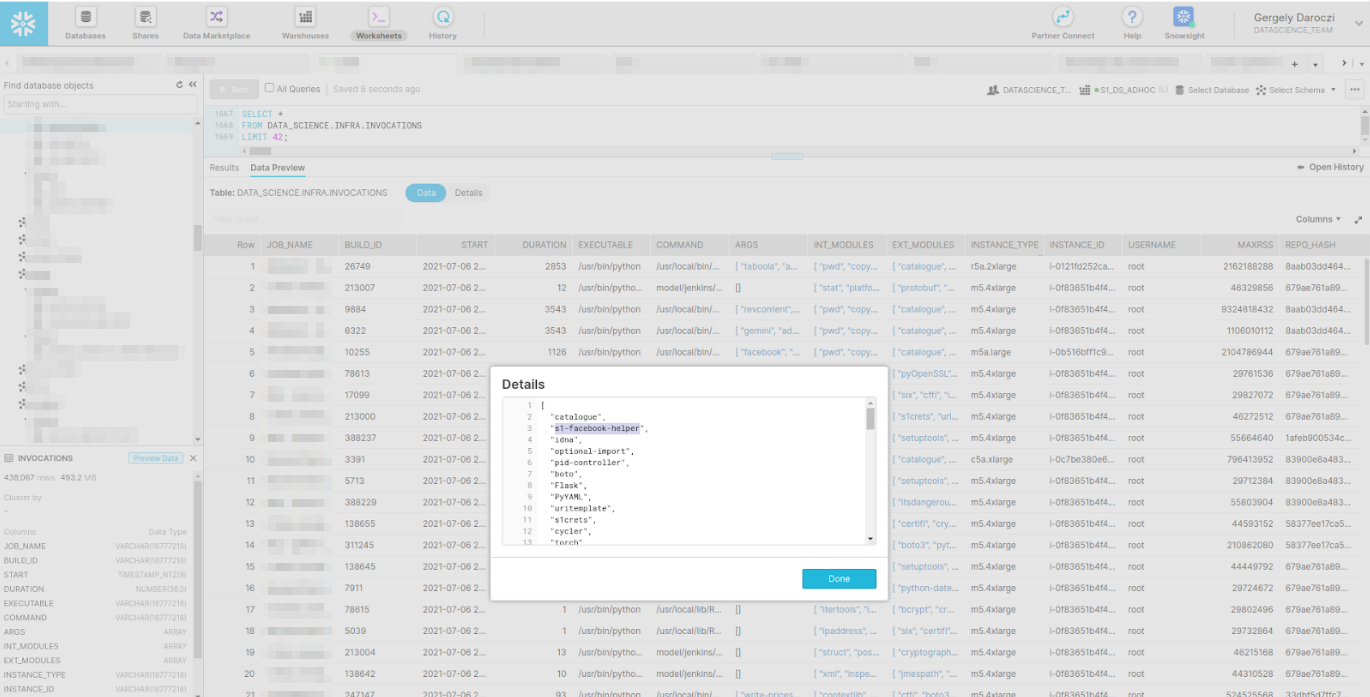
> ls(envir = ‘invocations’)
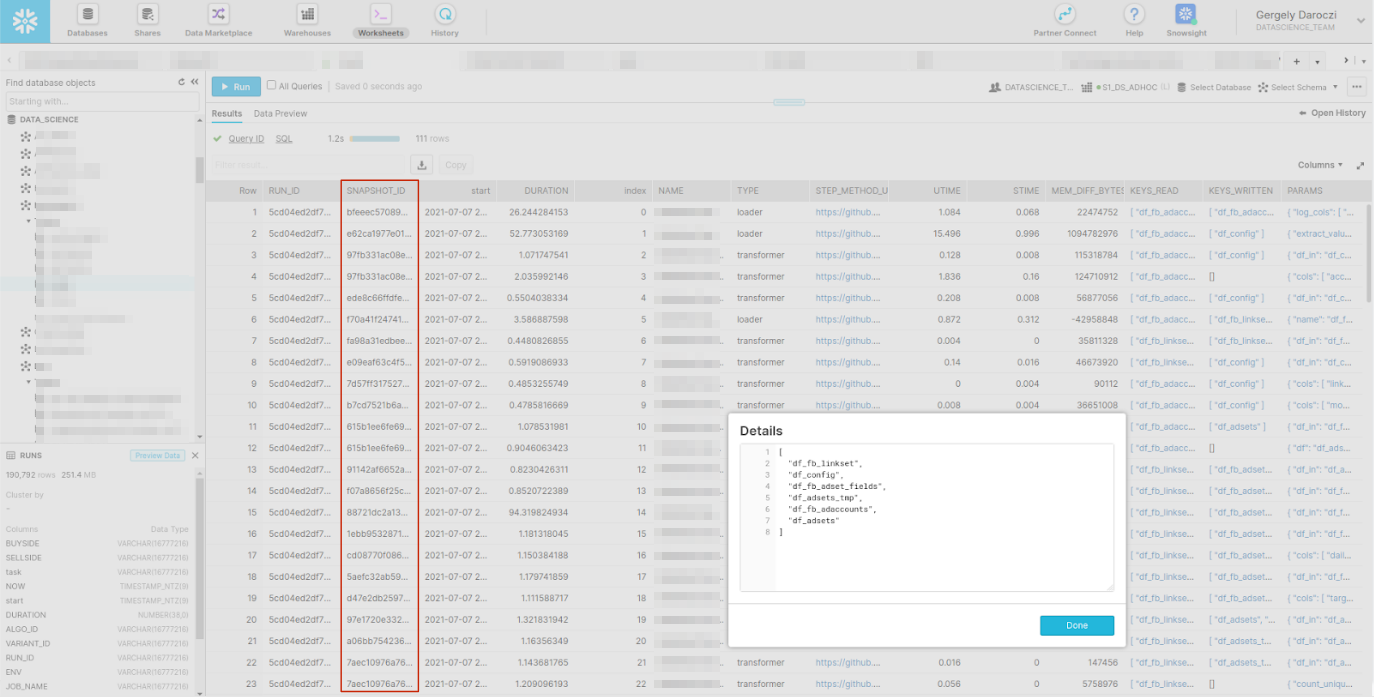
> ls(envir = ‘snapshots’)
> hire()
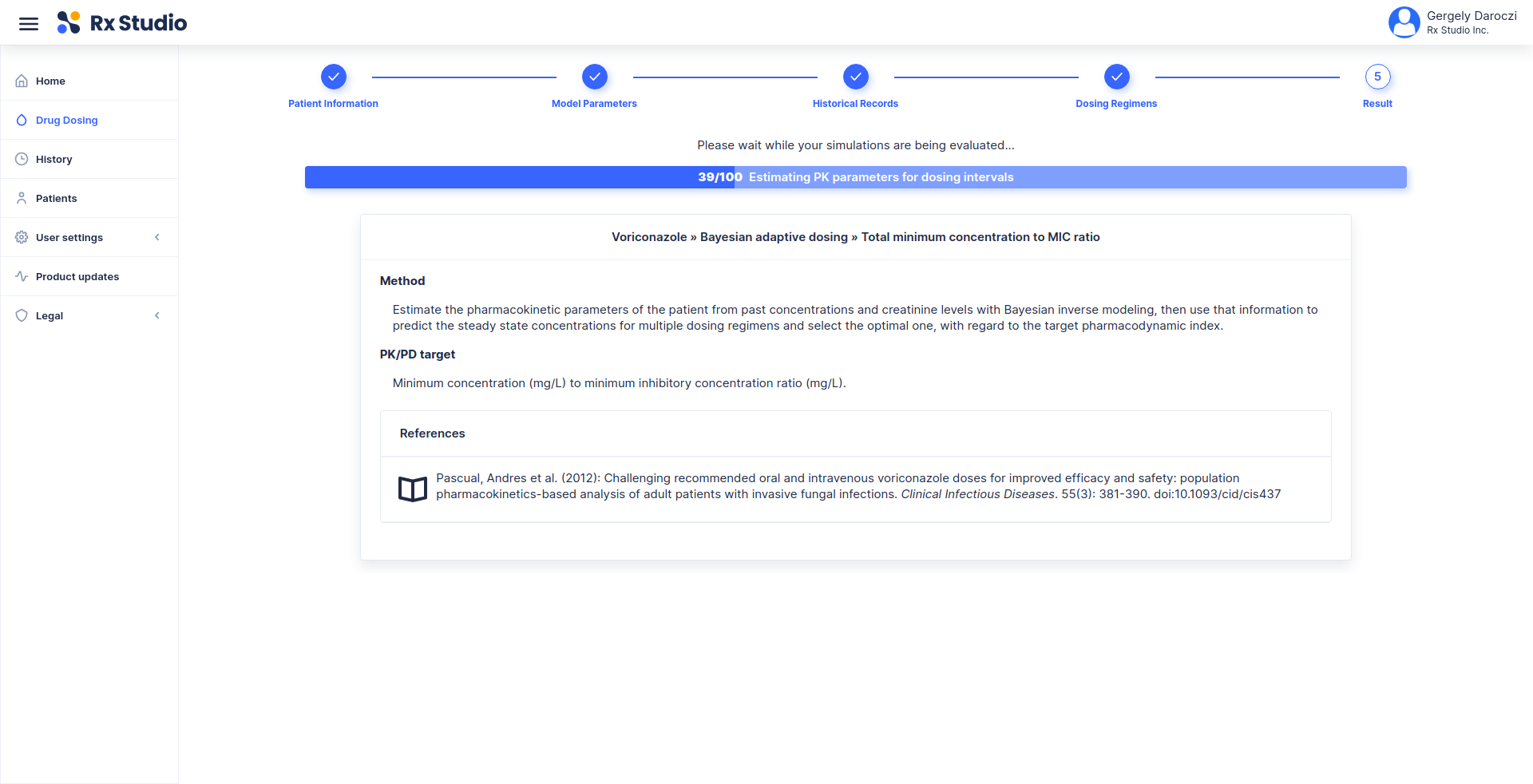
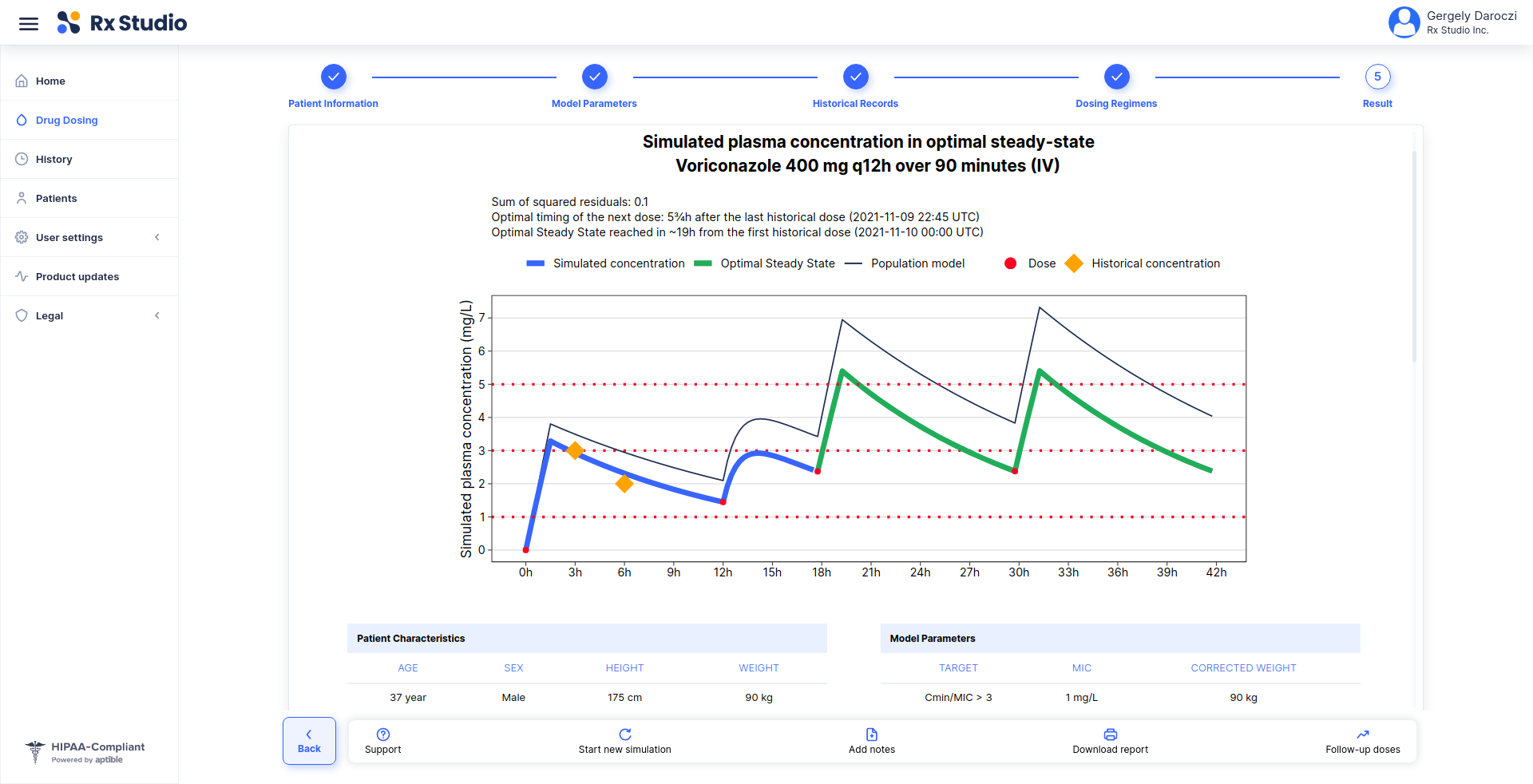
> demo(“rx.studio”)
> demo(“rx.studio”)
> demo(“rx.studio”)
> demo(“rx.studio”)
> demo(“rx.studio”)
> demo(“rx.studio”)
> demo(“rx.studio”)
> demo(“rx.studio”)
> demo(“rx.studio”)
> library(rx.studio)
<!--head
meta:
drug: ~
method: ~
target: ~
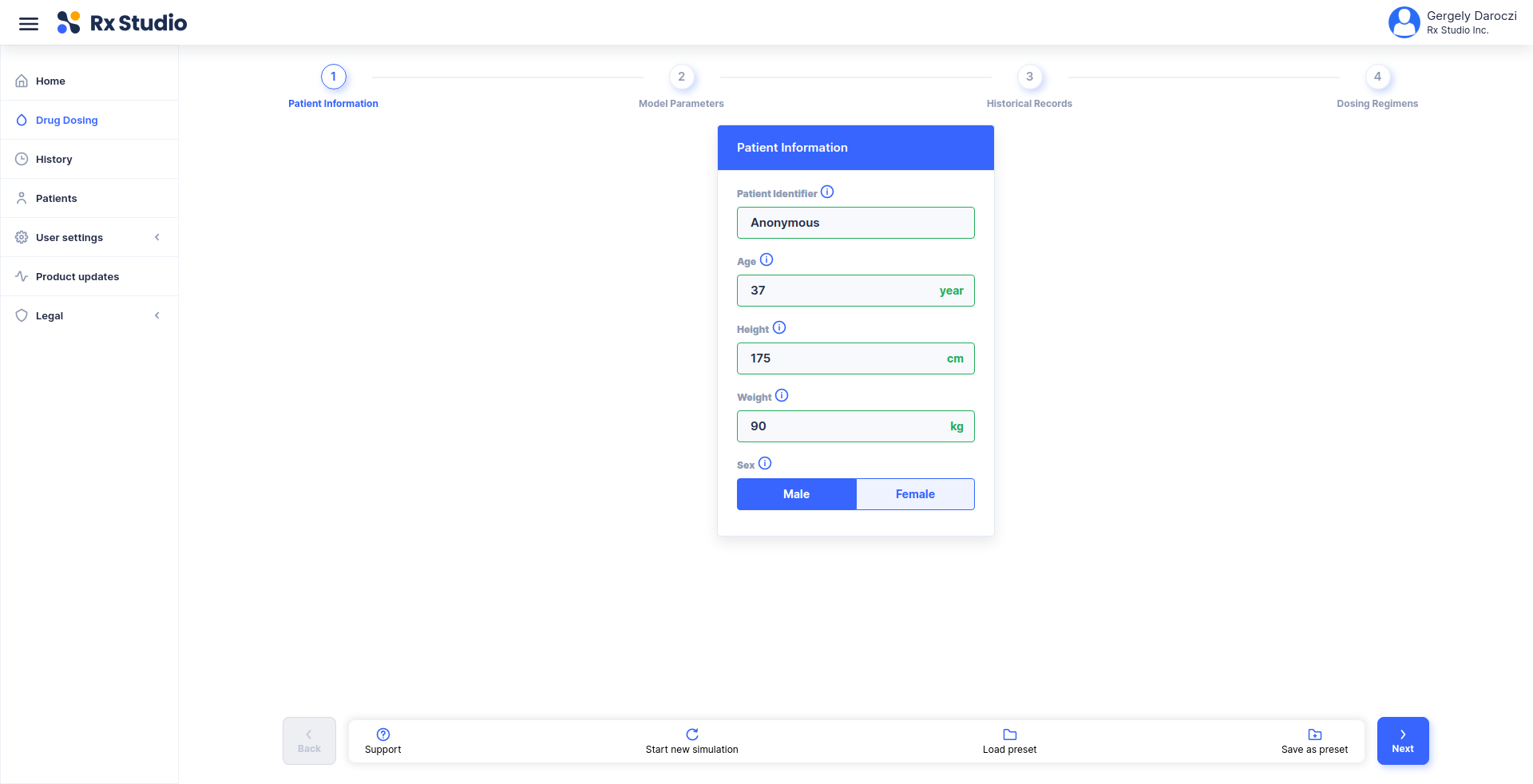
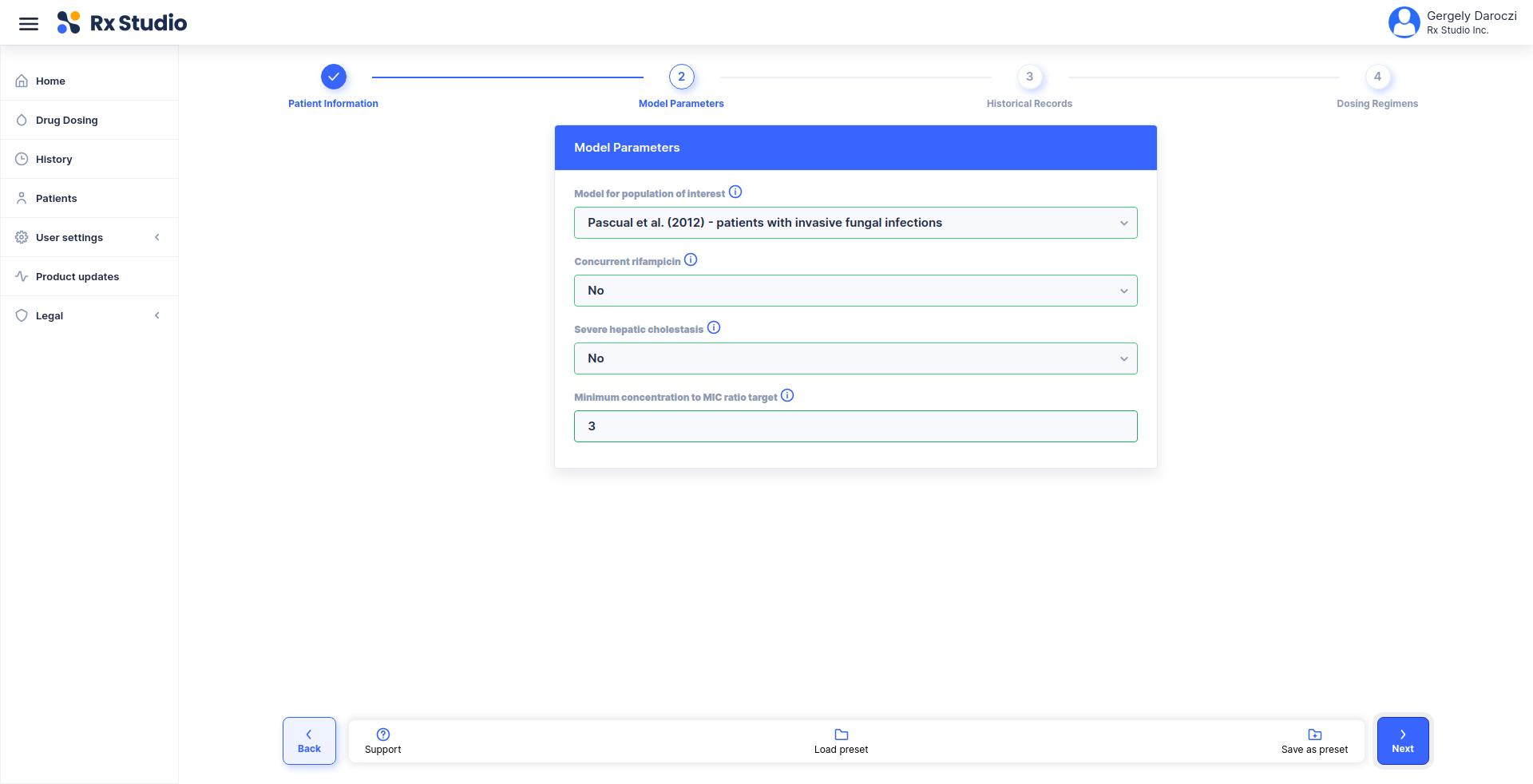
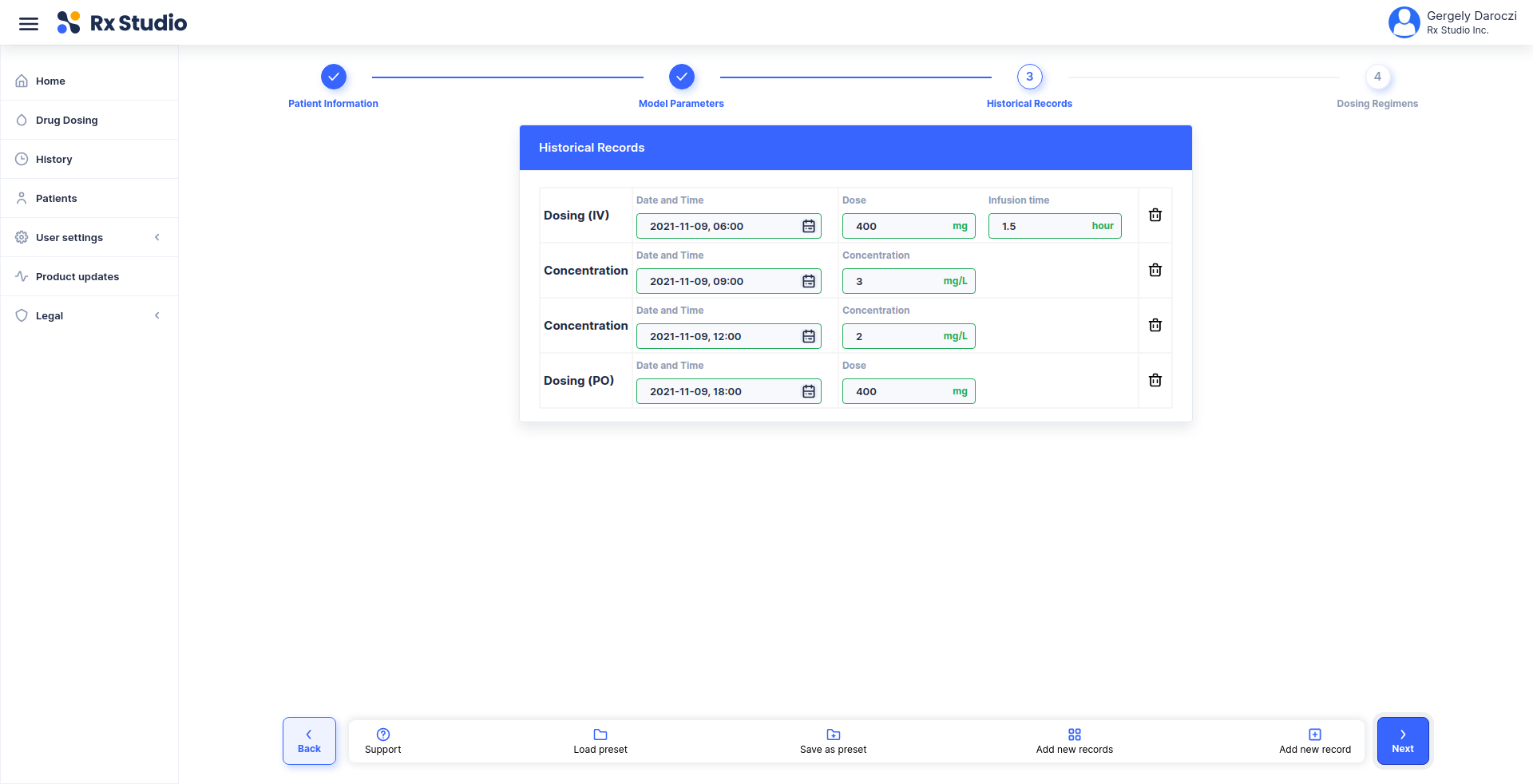
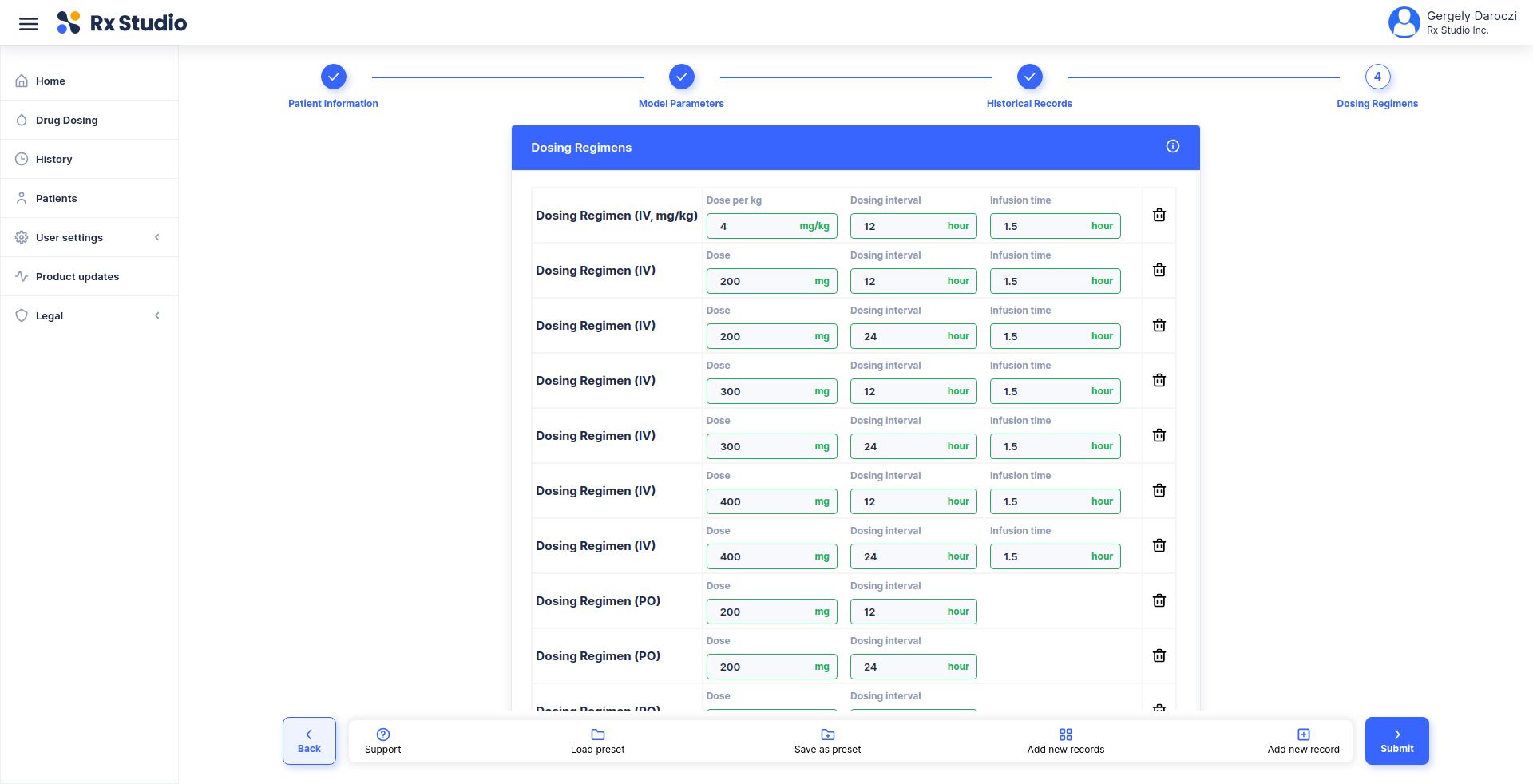
title: Calculate corrected weight for CrCl estimation
description: |
Using the Cockcroft-Gault 40% Obesity Adjustment for patients who are
greater than 30% of their ideal body weight.
packages:
- rx.studio
examples:
- list(HEIGHT = 174, WEIGHT = 72, SEX = 'Male')
inputs:
- !expr generate_input(type = 'HEIGHT')
- !expr generate_input(type = 'WEIGHT')
- !expr generate_input(type = 'SEX')
head-->
<%=
calc_cweight(HEIGHT, WEIGHT, SEX, adjthr = 1.3)
%>> library(rx.studio)
<!--head
meta:
drug: ~
method: ~
target: ~
title: Calculate corrected weight for CrCl estimation
description: |
Using the Cockcroft-Gault 40% Obesity Adjustment for patients who are
greater than 30% of their ideal body weight.
packages:
- rx.studio
examples:
- list(HEIGHT = 174, WEIGHT = 72, SEX = 'Male')
inputs:
- !expr generate_input(type = 'HEIGHT')
- !expr generate_input(type = 'WEIGHT')
- !expr generate_input(type = 'SEX')
head-->
<%=
calc_cweight(HEIGHT, WEIGHT, SEX, adjthr = 1.3)
%>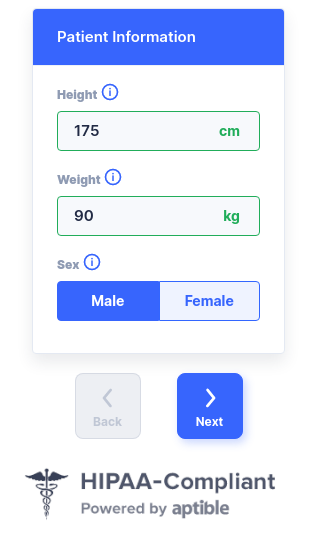
> str(“rx.studio”)
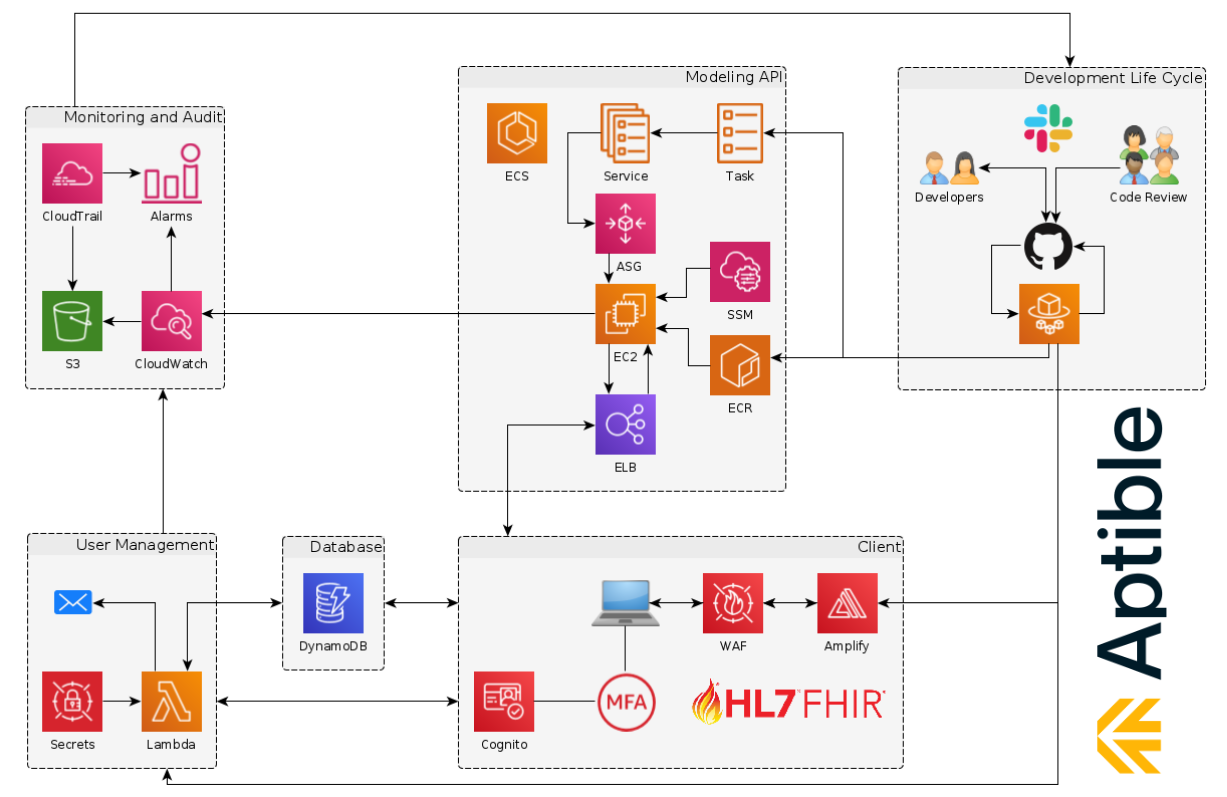
> is.compliant(“rx.studio”)

Source: the_coding_love – When the library has good documentation
> is.compliant(“rx.studio”)

> audit(“rx.studio”)
> audit(“rx.studio”)
> audit(“rx.studio”)
> audit(“rx.studio”)
> audit(“rx.studio”)
> audit(“rx.studio”)
> audit(“rx.studio”)
List of 4+
$ use_common_sense: TRUE
$ policies: function(...) search(...) |> get |> apply |> assert
$ data_management: List of 3+
..$ encrypt: List of 2
.. ..$ in_transit: TRUE
.. ..$ at_rest: TRUE
..$ document: TRUE
..$ PHI: identify()
.. ..
$ vendor_management: List of 3+
..$ cannot_live_without: TRUE
..$ security_assessment: TRUE
..$ BAA: TRUE
.. ..
...> audit(“rx.studio”)
List of 5+
$ use_common_sense: TRUE
$ policies: function(...) search(...) |> get |> apply |> assert
$ data_management: List of 3+
..$ encrypt: List of 2
.. ..$ in_transit: TRUE
.. ..$ at_rest: TRUE
..$ document: TRUE
..$ PHI: identify()
.. ..
$ vendor_management: List of 3+
..$ cannot_live_without: TRUE
..$ security_assessment: TRUE
..$ BAA: TRUE
.. ..
$ code_review: TRUE
...> audit(“rx.studio”)
List of Inf
$ use_common_sense: TRUE
$ policies: function(...) search(...) |> get |> apply |> assert
$ data_management: List of 3+
..$ encrypt: List of 2
.. ..$ in_transit: TRUE
.. ..$ at_rest: TRUE
..$ document: TRUE
..$ PHI: identify()
.. ..
$ vendor_management: List of 3+
..$ cannot_live_without: TRUE
..$ security_assessment: TRUE
..$ BAA: TRUE
.. ..
$ code_review: TRUE
$ unit_tests: TRUE
$ integration_tests: TRUE
$ code_coverage_tests: TRUE
...> readLines('frontend/es.po', n=25)
# Copyright (C) 2020-2021 Rx Studio Inc.
msgid ""
msgstr ""
"Project-Id-Version: rx.studio.webapp 1.0\n"
"POT-Creation-Date: 2020-12-06 00:40\n"
"PO-Revision-Date: 2021-05-26 01:40:46\n"
"Last-Translator: Rx Studio <support@rx.studio>\n"
"Language-Team: Rx Studio <support@rx.studio>\n"
"Language: es\n"
"MIME-Version: 1.0\n"
"Content-Type: text/plain; charset=UTF-8\n"
"Content-Transfer-Encoding: 8bit\n"
msgctxt "account_verify.new_password"
msgid "New Password"
msgstr "Contraseña nueva"
#. User status meaning the user has subscribed to product updates/email newsletter.
msgctxt "common.subscribed"
msgid "Subscribed"
msgstr "Suscrito"> readLines('backend/pt.po', n=25)
# Copyright (C) 2020-2021 Rx Studio Inc.
msgid ""
msgstr ""
"Project-Id-Version: rx.studio.models 1.0\n"
"POT-Creation-Date: 2020-12-06 00:40\n"
"PO-Revision-Date: 2021-06-22 03:45:21\n"
"Last-Translator: Rx Studio <support@rx.studio>\n"
"Language-Team: Rx Studio <support@rx.studio>\n"
"Language: pt\n"
"MIME-Version: 1.0\n"
"Content-Type: text/plain; charset=UTF-8\n"
"Content-Transfer-Encoding: 8bit\n"
msgid "Total maximum concentration"
msgstr "Concentração máxima total"
#. Drug name, only translate if has a local name or version in your language.
msgid "Cefepime"
msgstr "Cefepima"
#. The f prefix refers to free, so a shorthand for Free AUC to MIC ratio.
msgid "fAUC/MIC"
msgstr "fASC/CIM"> translations_read() |> summary()
$ pocount ~/projects/rx.studio-webapp/src/assets/i18n/po/es.po \
~/projects/rx.studio-models/inst/i18n/es.po
Type Strings Words (source) Words (translation)
Translated: 383 (100%) 2498 (100%) 2979
Untranslated: 0 ( 0%) 0 ( 0%) n/a
Total: 383 2498 2979
Type Strings Words (source) Words (translation)
Translated: 246 (100%) 1234 (100%) 1360
Untranslated: 0 ( 0%) 0 ( 0%) n/a
Total: 246 1234 1360
Processing file : TOTAL:
Type Strings Words (source) Words (translation)
Translated: 629 (100%) 3732 (100%) 4339
Untranslated: 0 ( 0%) 0 ( 0%) n/a
Total: 629 3732 4339
File count: 2> testthat::test_package()

Source: the_coding_love – When I want to commit and Jenkins is not OK with it