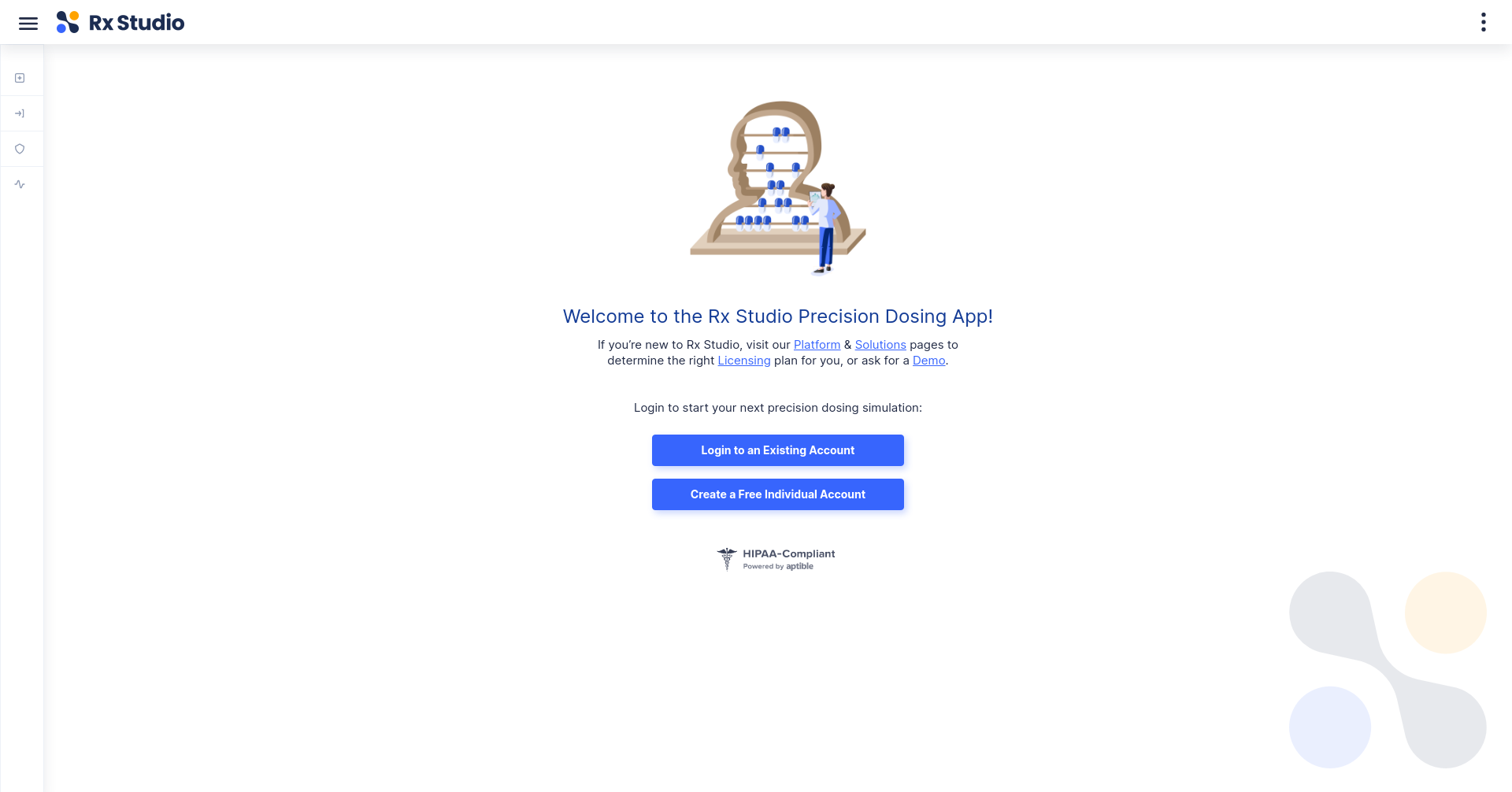
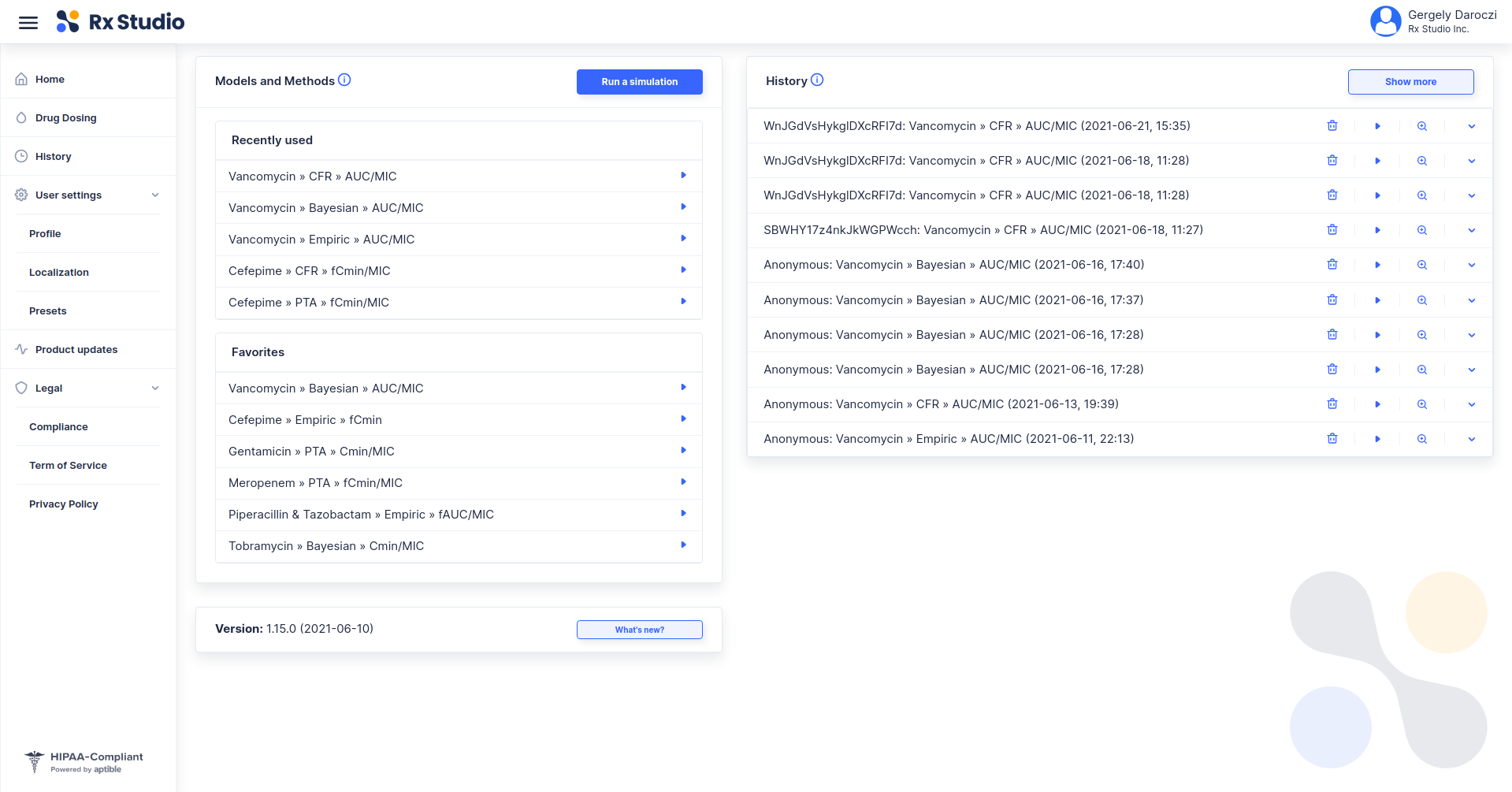
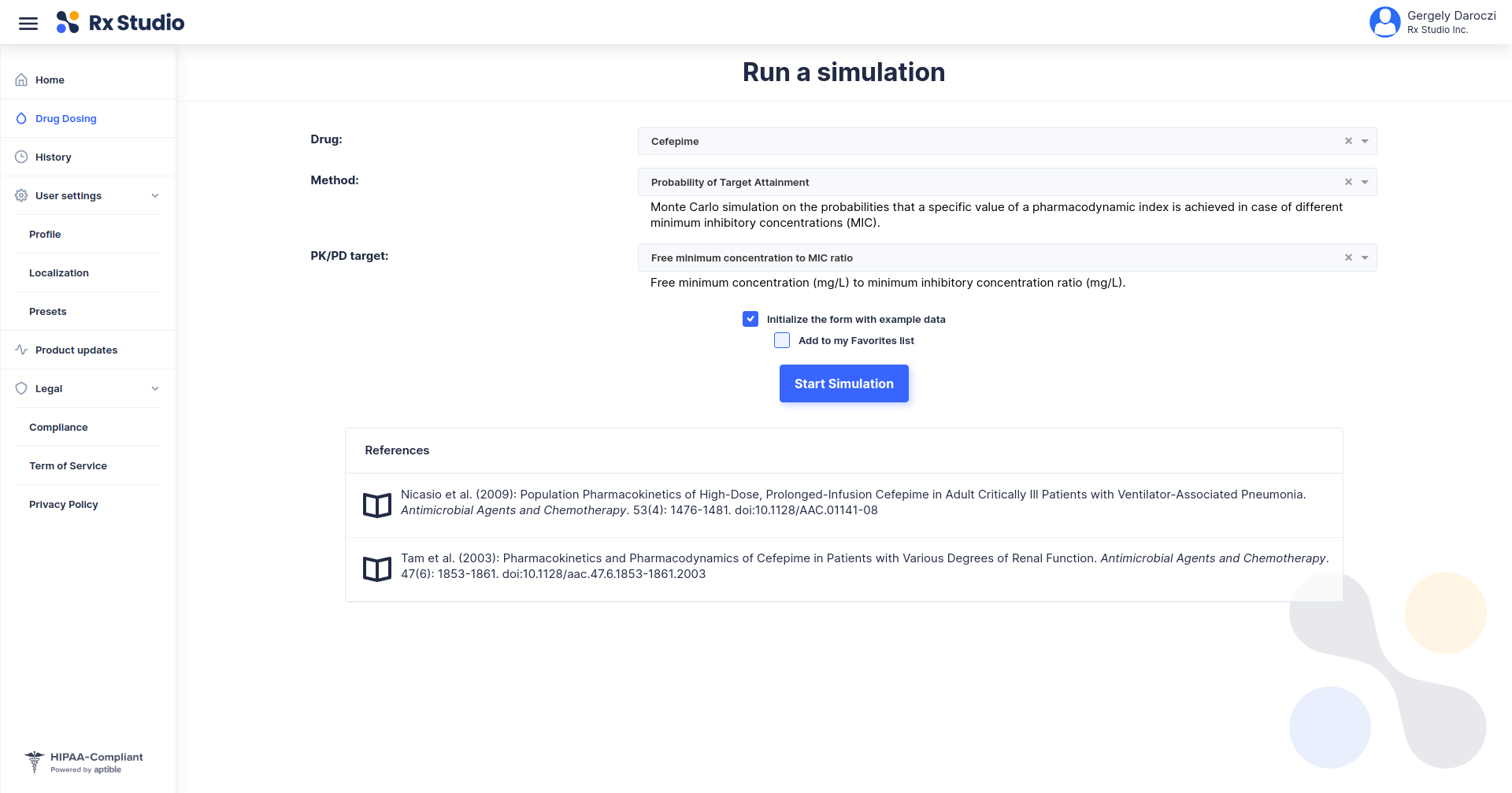
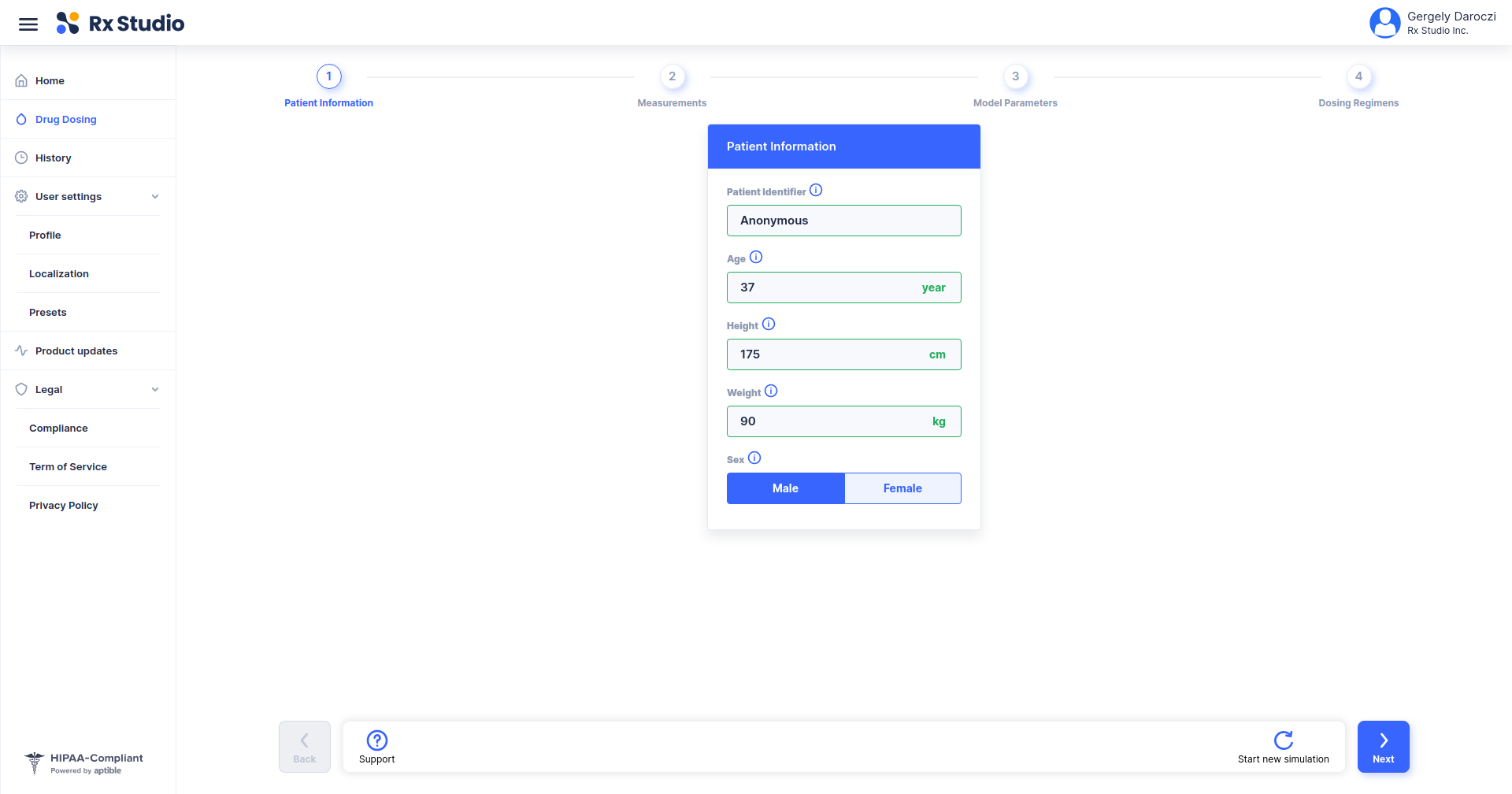
Using R to Empower a Precision Dosing Web Application
Gergely Daróczi
@daroczig
dummy slide
> whoami()
> whoami()
> whoami()
> whoami()
> whoami()
> whoami()
> whoarewe()
> demo()
> demo()
> demo()
> demo()
> demo()
> demo()
> demo()
> demo()
> demo()
> Sys.setenv(env = "prod")
> Sys.setenv(env = "prod")

> ??production
> ??production
- 2006: Calling R scripts from PHP (both reading from MySQL) to generate custom plots embedded in a homepage
- 2008: Automated/batch R scripts to generate thousands of pages of crosstables, ANOVA and plots from SPSS with
pdflatex - 2011: Ruby on Rails web application with
RApacheandpandocto report in plain English (NoSQL databases, scaling, security, central error tracking etc) - 2012: Plain RApache web application for NLP and network analysis
- 2015: Standardizing the data infrastructure of a fintech startup to use R for reporting, batch jobs, and stream processing
- 2017: Redesign, monitor and scale the DS infrastructure of an adtech startup for batch training and live scoring
> production <<- list(…)
Using in R in a non-interactive way:
- Running R without manual intervention (e.g. scheduled via CRON, triggered via upstream job trigger or API request)
- Need for a standard, e.g. containerized environment (pinned R and package versions, OS packages,
.Rprofileetc) - Security! (e.g. safeguarded production environment, encrypted credentials, aware of Little Bobby Tables, AppArmor etc)
- Job output is informative (logging), recorded (logging) and monitored (e.g.
errorhandler for ErrBit, CloudWatch logs or Splunk etc), alerts and notifications
> plot(production)
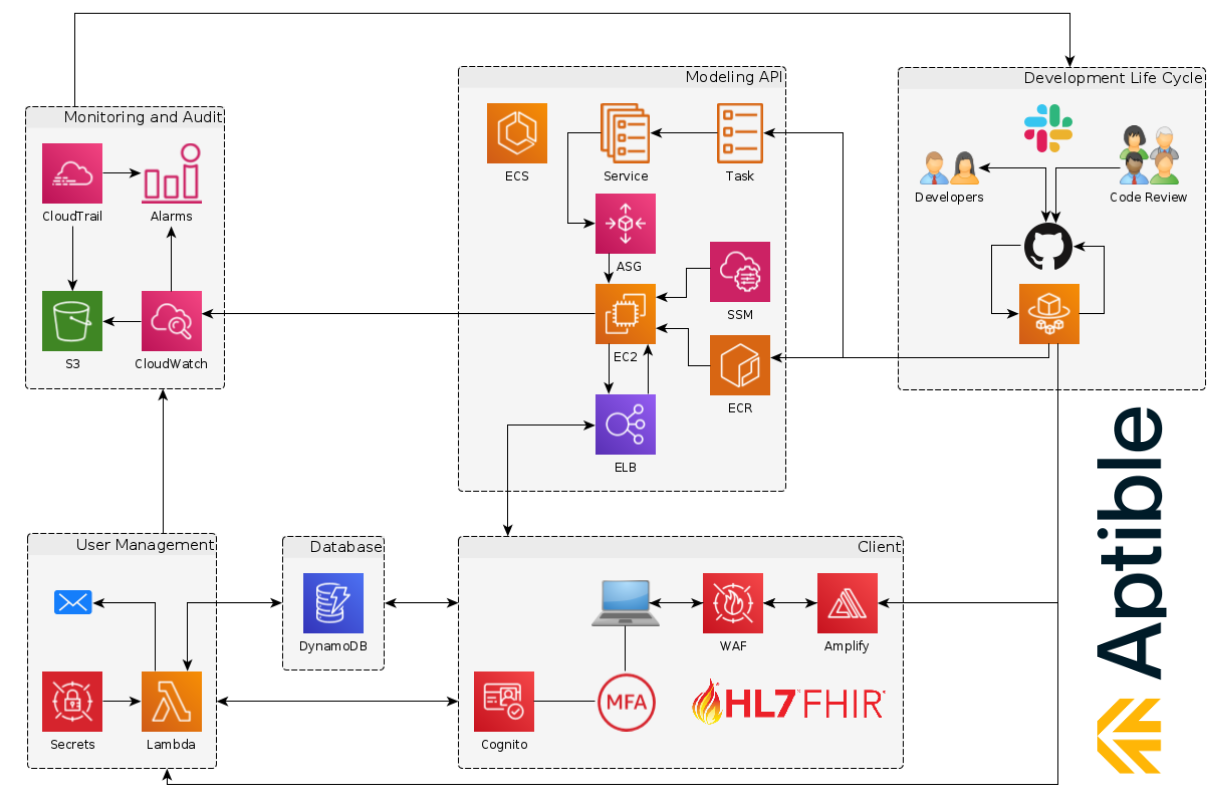
> is.compliant()
> is.compliant()

Source: the_coding_love – When the library has good documentation
> is.compliant()

> history()
> history()
- R Foundation – R: Regulatory Compliance and Validation Issues A Guidance Document for the Use of R in Regulated Clinical Trial Environments
Establishing documented evidence which provides a high degree of assurance that a specific process will consistently produce a product meeting its predetermined specifications and quality attributes. (FDA Glossary of Computer System Software Development Terminology)
> history()
- R Foundation – R: Regulatory Compliance and Validation Issues A Guidance Document for the Use of R in Regulated Clinical Trial Environments
Establishing documented evidence which provides a high degree of assurance that a specific process will consistently produce a product meeting its predetermined specifications and quality attributes. (FDA Glossary of Computer System Software Development Terminology)
- Mango Solutions – ValidR
> history()
- R Foundation – R: Regulatory Compliance and Validation Issues A Guidance Document for the Use of R in Regulated Clinical Trial Environments
Establishing documented evidence which provides a high degree of assurance that a specific process will consistently produce a product meeting its predetermined specifications and quality attributes. (FDA Glossary of Computer System Software Development Terminology)
- Mango Solutions – ValidR
- R Consortium – R Validation Hub
> str(audit)
> str(audit)
> str(audit)
> str(audit)
> str(audit)
> str(audit)
> str(audit)
List of 4+
$ use_common_sense: TRUE
$ policies: function(...) search(...) |> get |> apply |> assert
$ data_management: List of 3+
..$ encrypt: List of 2
.. ..$ in_transit: TRUE
.. ..$ at_rest: TRUE
..$ document: TRUE
..$ PHI: identify()
.. ..
$ vendor_management: List of 3+
..$ cannot_live_without: TRUE
..$ security_assessment: TRUE
..$ BAA: TRUE
.. ..
...> str(audit)
List of 5+
$ use_common_sense: TRUE
$ policies: function(...) search(...) |> get |> apply |> assert
$ data_management: List of 3+
..$ encrypt: List of 2
.. ..$ in_transit: TRUE
.. ..$ at_rest: TRUE
..$ document: TRUE
..$ PHI: identify()
.. ..
$ vendor_management: List of 3+
..$ cannot_live_without: TRUE
..$ security_assessment: TRUE
..$ BAA: TRUE
.. ..
$ code_review: TRUE
...> str(audit)
List of Inf
$ use_common_sense: TRUE
$ policies: function(...) search(...) |> get |> apply |> assert
$ data_management: List of 3+
..$ encrypt: List of 2
.. ..$ in_transit: TRUE
.. ..$ at_rest: TRUE
..$ document: TRUE
..$ PHI: identify()
.. ..
$ vendor_management: List of 3+
..$ cannot_live_without: TRUE
..$ security_assessment: TRUE
..$ BAA: TRUE
.. ..
$ code_review: TRUE
$ unit_tests: TRUE
$ integration_tests: TRUE
$ code_coverage_tests: TRUE
...
Source: twitter.com/romain_francois/status/1410886001539567616
> readLines('frontend/es.po', n=25)
# Copyright (C) 2020-2021 Rx Studio Inc.
msgid ""
msgstr ""
"Project-Id-Version: rx.studio.webapp 1.0\n"
"POT-Creation-Date: 2020-12-06 00:40\n"
"PO-Revision-Date: 2021-05-26 01:40:46\n"
"Last-Translator: Rx Studio <support@rx.studio>\n"
"Language-Team: Rx Studio <support@rx.studio>\n"
"Language: es\n"
"MIME-Version: 1.0\n"
"Content-Type: text/plain; charset=UTF-8\n"
"Content-Transfer-Encoding: 8bit\n"
msgctxt "account_verify.new_password"
msgid "New Password"
msgstr "Contraseña nueva"
#. User status meaning the user has subscribed to product updates/email newsletter.
msgctxt "common.subscribed"
msgid "Subscribed"
msgstr "Suscrito"> readLines('backend/pt.po', n=25)
# Copyright (C) 2020-2021 Rx Studio Inc.
msgid ""
msgstr ""
"Project-Id-Version: rx.studio.models 1.0\n"
"POT-Creation-Date: 2020-12-06 00:40\n"
"PO-Revision-Date: 2021-06-22 03:45:21\n"
"Last-Translator: Rx Studio <support@rx.studio>\n"
"Language-Team: Rx Studio <support@rx.studio>\n"
"Language: pt\n"
"MIME-Version: 1.0\n"
"Content-Type: text/plain; charset=UTF-8\n"
"Content-Transfer-Encoding: 8bit\n"
msgid "Total maximum concentration"
msgstr "Concentração máxima total"
#. Drug name, only translate if has a local name or version in your language.
msgid "Cefepime"
msgstr "Cefepima"
#. The f prefix refers to free, so a shorthand for Free AUC to MIC ratio.
msgid "fAUC/MIC"
msgstr "fASC/CIM"> translate
function(text, language = translate_get_language()) {
if (!language %in% translation_languages()) {
stop(paste('Unknown language:', shQuote(language)))
}
if (language == 'en') {
translation <- text
} else {
translation <- translations[[language]][[text, exact = TRUE]]
}
if (is.null(translation) || translation == '') {
stop(paste('Missing translation of', shQuote(text),
'in language', shQuote(language)))
}
translation
}
<environment: namespace:rx.studio>> translation_read
function(language, folder = system.file('i18n', package = 'rx.studio')) {
po <- readLines(file.path(folder, paste(language, 'po', sep = '.')))
getlines <- function(prefix) {
sub(paste0('^', prefix, ' "(.*)"$'), '\\1',
grep(paste0('^', prefix, ' '), po, value = TRUE))
}
msgids <- getlines('msgid')[-1]
msgstrs <- getlines('msgstr')[-1]
if (language == 'en') {
msgstrs <- msgids
}
as.list(setNames(msgstrs, msgids))
}
<environment: namespace:rx.studio>> .onLoad <- function(lib, pkg) {
translations <- list2env(list(default = 'en'), parent = emptyenv())
#' List supported translations
#' @return character vector of language codes
#' @export
translation_languages <- function() {
files <- list.files(system.file('i18n', package = 'rx.studio'), pattern = '.po$')
assign('languages', sub('.po', '', basename(files)), envir = translations)
translations$languages
}
#' Cache all translations
#' @return variables in the \code{translations} environment of the package
#' @export
translations_read <- function() {
for (language in translation_languages()) {
assign(language,
as.environment(translation_read(language)),
envir = translations)
}
}> translations_generate
## based on tools::xgettext
find_strings <- function(e) {
if (is.call(e) && is.name(e[[1L]]) && (as.character(e[[1L]]) == 'translatable')) {
## extract text to be translated and comment to be passed to the translator
text <- eval(e[[2]])
if (!is.null(text) && test_string(text)) {
log_trace(skip_formatter(paste('Text to translate:', as.character(text))))
comment <- ifelse(length(e) > 2, eval(e[[3]]), NA_character_)
strings <<- c(strings, list(list(text = text, comment = comment)))
}
} else {
if (is.recursive(e)) {
for (i in seq_along(e)) {
Recall(e[[i]])
}
}
}
}
## extract terms from package source, templats, reports etc.
lapply(list.files(pattern = '.R$', full.names = TRUE), find_strings)> translations_read() |> summary()
$ pocount ~/projects/rx.studio-webapp/src/assets/i18n/po/es.po \
~/projects/rx.studio-models/inst/i18n/es.po
Type Strings Words (source) Words (translation)
Translated: 383 (100%) 2498 (100%) 2979
Untranslated: 0 ( 0%) 0 ( 0%) n/a
Total: 383 2498 2979
Type Strings Words (source) Words (translation)
Translated: 246 (100%) 1234 (100%) 1360
Untranslated: 0 ( 0%) 0 ( 0%) n/a
Total: 246 1234 1360
Processing file : TOTAL:
Type Strings Words (source) Words (translation)
Translated: 629 (100%) 3732 (100%) 4339
Untranslated: 0 ( 0%) 0 ( 0%) n/a
Total: 629 3732 4339
File count: 2> testthat::test_package()

Source: the_coding_love – When I want to commit and Jenkins is not OK with it
> testthat::test_package()
on: pull_request
name: Repository tools
jobs:
all-translated:
runs-on: ubuntu-latest
steps:
- uses: actions/checkout@v2
- uses: aws-actions/configure-aws-credentials@v1
- uses: aws-actions/amazon-ecr-login@v1
- name: Pull prod Docker image from Amazon ECR
run: docker pull $ECR_REGISTRY/r-api-base
- name: Build test Docker image locally
run: docker build --file inst/docker/r-api/Dockerfile -t test .
- name: all translation terms extracted
run: library(rx.studio); assert_translatable_extracted('/R')
shell: docker run --rm --init -v /home:/home -w /R test Rscript {0}
- name: all English terms are translated
run: quit(save='no', status=sum(as.data.frame(testthat::test_file('inst/tests/testthat/test-translate.R'))$failed))
shell: docker run --rm --init -v /home:/home -w /R test Rscript {0}> sessionInfo()


> library(logger)
library(logger)
log_threshold(DEBUG)
log_info('Script starting up...')
#> INFO [2018-20-11 22:49:36] Script starting up...
pkgs <- available.packages()
log_info('There are {nrow(pkgs)} R packages hosted on CRAN!')
#> INFO [2018-20-11 22:49:37] There are 13433 R packages hosted on CRAN!
for (letter in letters) {
lpkgs <- sum(grepl(letter, pkgs[, 'Package'], ignore.case = TRUE))
log_level(if (lpkgs < 5000) TRACE else DEBUG,
'{lpkgs} R packages including the {shQuote(letter)} letter')
}
#> DEBUG [2018-20-11 22:49:38] 6300 R packages including the 'a' letter
#> DEBUG [2018-20-11 22:49:38] 6772 R packages including the 'e' letter
#> DEBUG [2018-20-11 22:49:38] 5412 R packages including the 'i' letter
#> DEBUG [2018-20-11 22:49:38] 7014 R packages including the 'r' letter
#> DEBUG [2018-20-11 22:49:38] 6402 R packages including the 's' letter
#> DEBUG [2018-20-11 22:49:38] 5864 R packages including the 't' letter> library(logger)
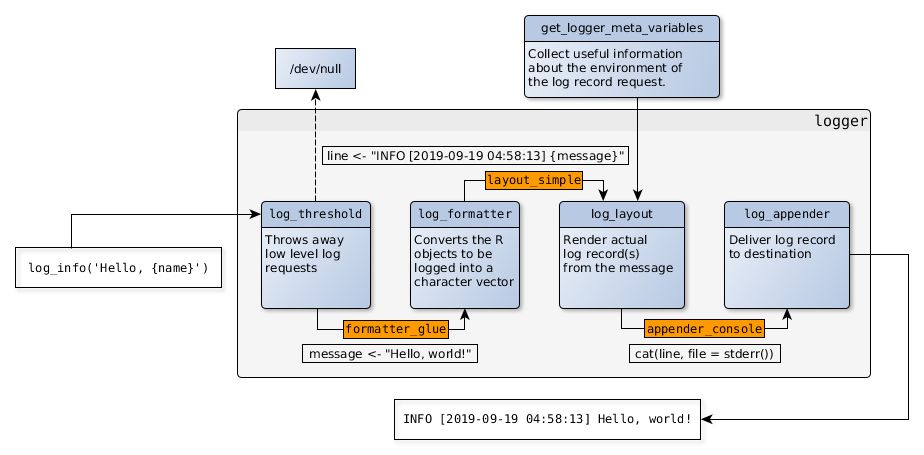
> library(logger)
> library(logger)

> news()
Source: the_coding_love – When I receive a pull request with dozens of commits
> news()
#' List of changes affecting simulation outputs
#'
#' Each item is a \code{list} with the below required fields:
#' \itemize{
#' \item \code{release} semver change that can be \code{init}, \code{patch}, \code{minor} or \code{major}
#' \item \code{date} date of change
#' \item \code{scope} list of affected drugs, methods and targets
#' \item \code{description} One-line description of the change.
#' }
#' @keywords internal
CHANGELOG <- list(
list(
release = 'init',
date = as.Date('2020-09-05'),
scope = list(drugs = c('cefepime'), methods = c('cfr', 'pta', 'emp', 'bay'), targets = c('fcmin_mic_ratio', 'ftime_above_mic'))),
list(
release = 'major',
date = as.Date('2020-09-07'),
scope = list(drugs = c('cefepime'), methods = c('pta'), targets = c('fcmin_mic_ratio', 'ftime_above_mic')),
description = 'Enable multiple dosing regimens.'),
...> news()
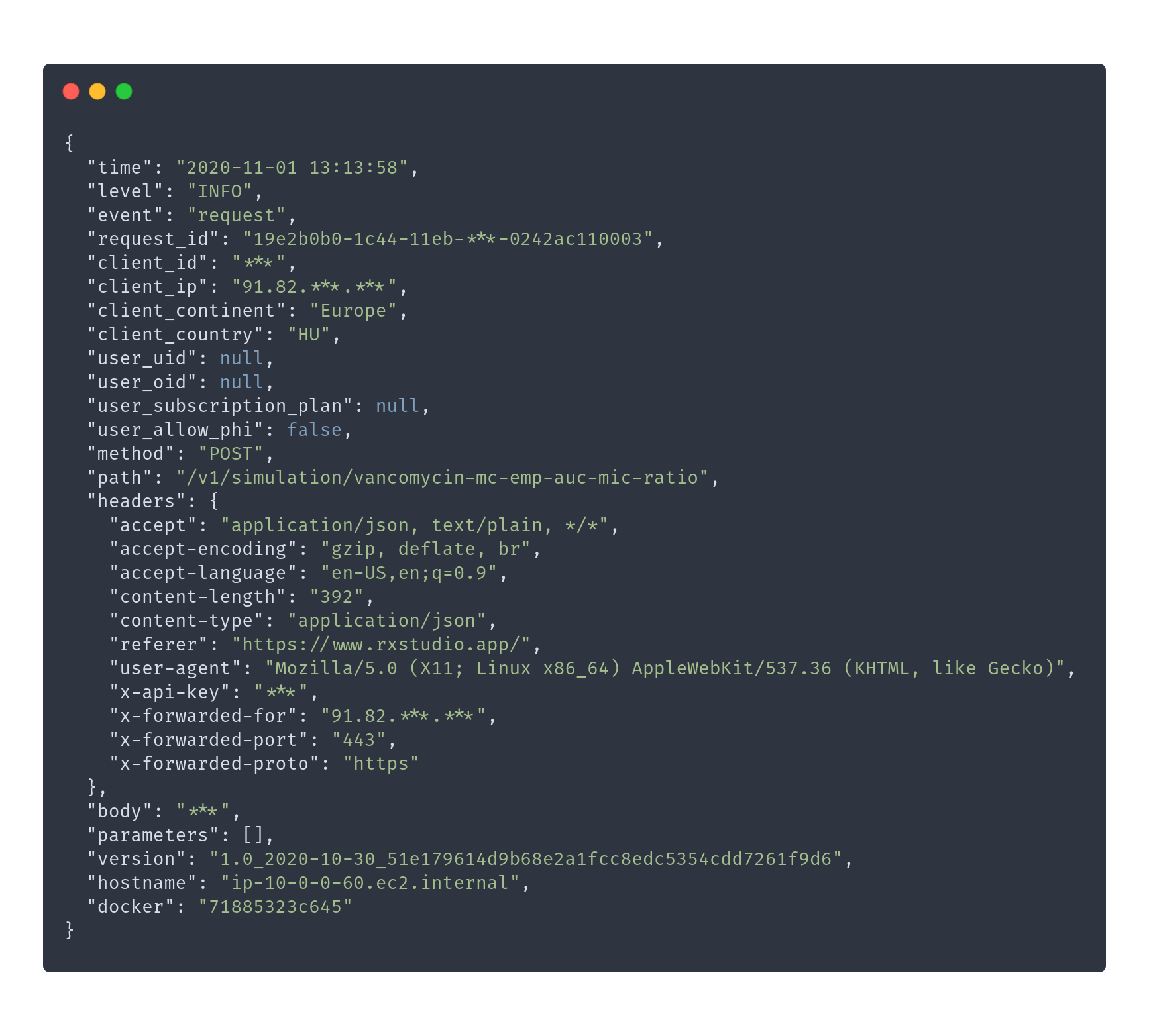
> do.call(evaluate_simulation, …)
#* @filter language
function(res, req) {
log_trace(step = 'Start of language filter')
req$language <- req$HEADERS['accept-language']
if (is.na(req$language)) {
req$language <- 'en'
}
tryCatch(
translate_set_language(req$language),
error = function(e) {
log_error(
event = 'invalid language',
request_id = req$request_id,
language = req$language)
}
)
log_trace(step = 'End of language filter')
plumber::forward()
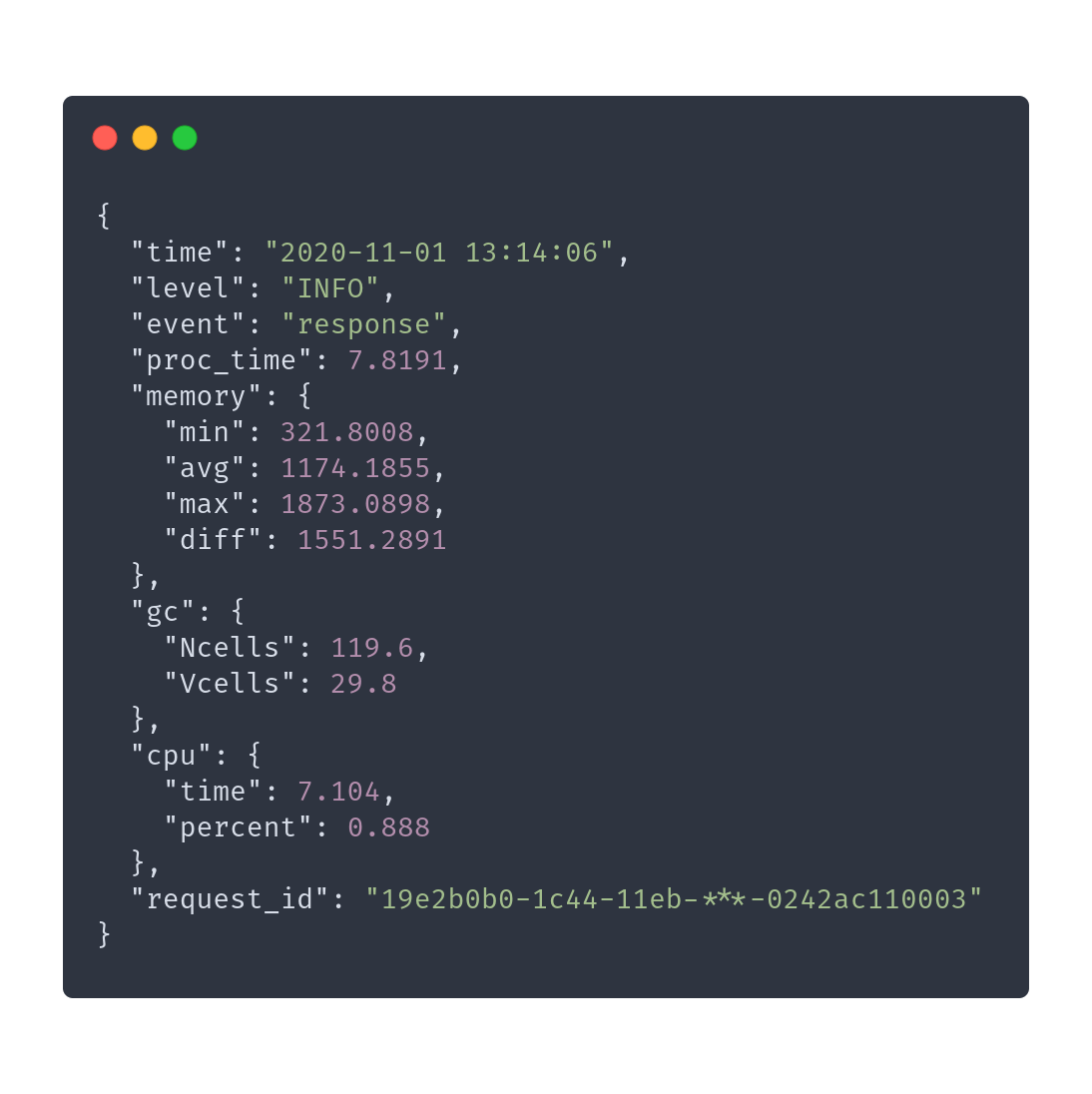
}> do.call(evaluate_simulation, …)
# ...
id_token <- sub('^Bearer ', '', req$HTTP_AUTHORIZATION)
firebase_admin <- reticulate::import(module = 'firebase_admin')
app <- firebase_admin$initialize_app()
on.exit(firebase_admin$delete_app(app))
user <- firebase_admin$auth$verify_id_token(id_token)
log_debug(
event = 'authenticated',
client_id = req$client_id,
user_uid = user$user_uid,
user_oid = user$user_oid,
user_subscription_plan = user$user_subscription_plan,
user_allow_phi = user$user_allow_phi,
language = headers[['accept-language']])
# ...> do.call(evaluate_simulation, …)
template <- read_template(path)
env <- validate_template_inputs(template, ...)
## capture stderr in forked process
fork_std_err_con <- rawConnection(raw(0), 'r+')
on.exit(try(close(fork_std_err_con), silent = TRUE), add = TRUE)
log_trace(step = 'Evaluating template')
report <- tryCatch(suppressMessages(suppressWarnings(
eval_safe(
Pandoc.brew(text = template$body, envir = env, output = '/dev/null'),
std_err = fork_std_err_con))),
error = function(e) {
msg <- paste(rawToChar(rawConnectionValue(fork_std_err_con)), collapse = '')
if (length(msg) == 0 || msg == '') {
msg <- e$message
}
stop(msg)
})
s3_upload_file(tarfile, file.path(CONSTANTS$infra$s3$buckets$simulations, env$request_id))> read_template('vanco-bay-auc-mic')
<!--head
meta:
drug: !ref vancomycin
method: !ref bayesian
target: !ref auc_mic_ratio
references: !ref
- goti_2018
- soetaert_2010
- lodise_2009
- buelga_2005
- rybak_2020
inputs:
- !expr generate_input(type = 'PATID')
- !expr generate_input(type = 'AGE')
- !expr generate_input(type = 'MODEL',
labels = c(
translatable('Goti et al. (2018) - Patients NOT undergoing hemodialysis'),
translatable('Goti et al. (2018) - Patients undergoing hemodialysis'),
translatable('Buelga et al. (2005) - Patients with hematological malignancies'),
translatable('Buelga et al. (2005) - AML patients, Model 1'),
translatable('Buelga et al. (2005) - AML patients, Model 2')),
default = 'Goti et al. (2018) - Patients NOT undergoing hemodialysis')
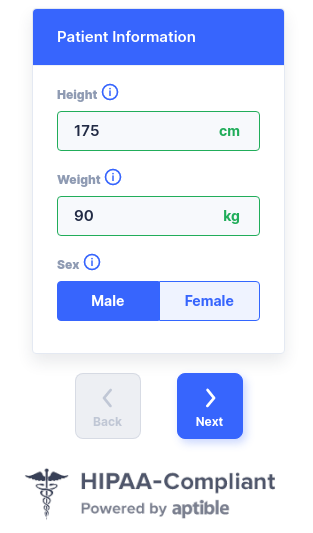
...> generate_input(type = 'PATID')
> generate_input(type = 'MODEL')
> ??assert_factor_level
#' Make it easy to reference a factor's possible level and fail on not existing label
#' @export
#' @examples
#' MODEL <- factor(levels = c('ICU', 'General Ward'))
#'
#' MODEL$ICU
#' # [1] ICU
#'
#' MODEL$foobar
#' # Error: Unkonwn category of 'MODEL', should be one of 'ICU', 'General Ward'
`$.factor` <- function(object, key) {
if (!key %in% levels(object)) {
stop(sprintf(
'Unknown category of %s, should be one of %s',
shQuote(deparse(substitute(object))),
paste(shQuote(levels(object)), collapse = ', ')
))
}
key
}> read_template('vanco-bay-auc-mic')
<% ## Process data
z <- initialize_model_parameters()
z <- systematize_historical_data(z)
z <- assess_patient_parameters(z)
z <- simulate_pk_parameters(z)
z <- run_calculations(z)
%>
<% ## Output %>
<% for (id in z$output_ids) { %>
<%= display_results(z, id) %>
<% } %>
<%= log_results(z) %>> CONSTANTS
list2env(methods = list(
'bayesian' = list(
id = 'bay',
title = translatable( 'Bayesian adaptive dosing', 'OK to leave untranslated if there is no standard translation in the local scientific language.'), # nolint
description = translatable('Estimate the pharmacokinetic parameters of the patient from past concentrations and creatinine levels with Bayesian inverse modeling, then use that information to predict the steady state concentrations for multiple dosing regimens and select the optimal one, with regard to the target pharmacodynamic index.', 'Helper text shown in web app to users describing what the Bayesian adaptive dosing method does.'), # nolint
output_ids = c(
'plot_simconc_ss',
'excerpt_separator',
'display_messages',
'table_patient_params',
'table_model_params',
'plot_target_param_ss',
'plot_regimens_aucpermic',
'plot_regimens_pctabovemic',
'plot_regimens_cmin_cmax',
'plot_regimens_toxicity',
'table_pk_params'
)
)
)> ??calc
#' Calculate Lean Body Weight (LBW)
#'
#' @param sex SEX input
#' @inheritParams calc_bmi
#' @return LBW (kg)
#' @references Janmahasatian, Sarayut et al. "Quantification of lean bodyweight."
#' Clinical pharmacokinetics vol. 44,10 (2005): 1051-65. \url{https://doi.org/10.2165/00003088-200544100-00004}
#' @export
#' @importFrom data.table fcase
calc_lbw <- function(sex, height, weight) {
# check args
assert_sex(sex)
# main
mweight <- 9270 * weight
bmi <- calc_bmi(height, weight)
# return
fcase(
sex == sex$Male, mweight / (6680 + 216 * bmi),
sex == sex$Female, mweight / (8780 + 244 * bmi)
)
}> ??calc
#' Calculate 24 hour AUC (via trapezoidal rule), Cmin (as last concentration) and Cmax
#' for a single dosing interval (i.e. a single dose at the beginning of the interval)
#'
#' @param d data.table with concentration data
#' @return data.table with AUC, Cmin & Cmax
#' @export
#' @importFrom checkmate assert_data_table assert_names
#' @importFrom data.table data.table := .N
#' @examples
#' calc_auc_cmin_cmax(data.table::data.table(N = c(rep(1, 10), rep(2, 10)), y = 1:20))
calc_auc_cmin_cmax <- function(d) {
# silence "no visible global function/variable definition" R CMD check
`.` <- y <- N <- NULL
# check args
assert_data_table(d, any.missing = FALSE)
dvars <- c("N", "y")
assert_names(names(d), must.include = dvars)
# return
d[, .(auc = (sum(y) - (y[1] + y[.N]) / 2) * 24 / max(.N - 1, 1), cmin = y[.N], cmax = max(y)), by = N]
}> licence()
> contributors()

> library(rx.studio)
<!--head
meta:
drug: ~
method: ~
target: ~
title: Calculate corrected weight for CrCl estimation
description: |
Using the Cockcroft-Gault 40% Obesity Adjustment for patients who are
greater than 30% of their ideal body weight.
packages:
- rx.studio
examples:
- list(HEIGHT = 174, WEIGHT = 72, SEX = 'Male')
inputs:
- !expr generate_input(type = 'HEIGHT')
- !expr generate_input(type = 'WEIGHT')
- !expr generate_input(type = 'SEX')
head-->
<%=
calc_cweight(HEIGHT, WEIGHT, SEX, adjthr = 1.3)
%>> library(rx.studio)
<!--head
meta:
drug: ~
method: ~
target: ~
title: Calculate corrected weight for CrCl estimation
description: |
Using the Cockcroft-Gault 40% Obesity Adjustment for patients who are
greater than 30% of their ideal body weight.
packages:
- rx.studio
examples:
- list(HEIGHT = 174, WEIGHT = 72, SEX = 'Male')
inputs:
- !expr generate_input(type = 'HEIGHT')
- !expr generate_input(type = 'WEIGHT')
- !expr generate_input(type = 'SEX')
head-->
<%=
calc_cweight(HEIGHT, WEIGHT, SEX, adjthr = 1.3)
%>